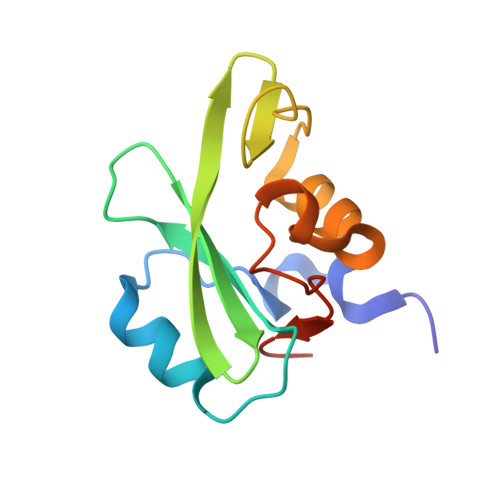
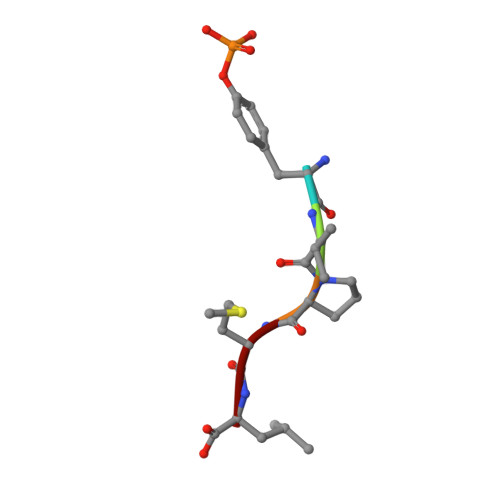
NMR Trial Models: Experiences with the Colicin Immunity Protein Im7 and the P85Alpha C-Terminal Sh2-Peptide Complex
Pauptit, R.A., Dennis, C.A., Derbyshire, D.J., Breeze, A.L., Weston, S.A., Rowsell, S., Murshudov, G.N.(2001) Acta Crystallogr D Biol Crystallogr 57: 1397
- PubMed: 11567151
- DOI: https://doi.org/10.1107/s0907444901012434
- Primary Citation of Related Structures:
1H9O - PubMed Abstract:
Two cases of successful molecular replacement using NMR trial models are presented. One is the crystal structure of the Escherichia coli colicin immunity protein Im7; the other is a heretofore unreported crystal structure of a specific PDGF receptor-derived peptide complex of the carboxy-terminal SH2 domain from the p85alpha subunit of human phosphatidylinositol 3-OH kinase. In both cases, molecular replacement was non-trivial. Success was achieved using trial models that consisted of an ensemble of NMR structures from which the more flexible portions had been excised. Use of maximum-likelihood refinement proved critical to be able to refine the poor starting models. The challenges typical of the use of NMR trial models in molecular replacement are discussed.
- AstraZeneca, Mereside, Alderley Park, Macclesfield, Cheshire SK10 4TG, England. richard.pauptit@astrazeneca.com
Organizational Affiliation: