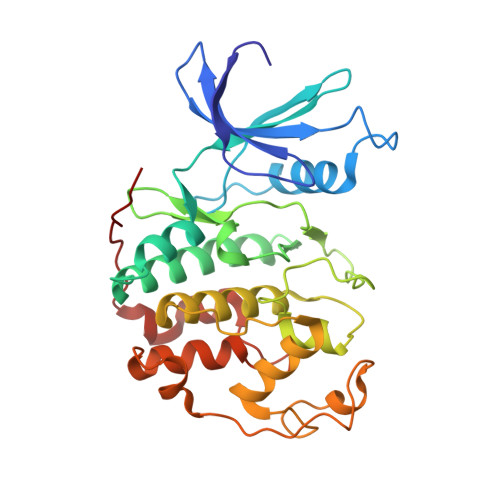
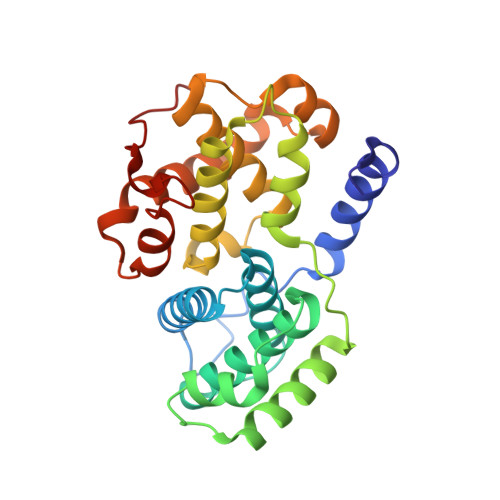
Prevention of chemotherapy-induced alopecia in rats by CDK inhibitors.
Davis, S.T., Benson, B.G., Bramson, H.N., Chapman, D.E., Dickerson, S.H., Dold, K.M., Eberwein, D.J., Edelstein, M., Frye, S.V., Gampe Jr, R.T., Griffin, R.J., Harris, P.A., Hassell, A.M., Holmes, W.D., Hunter, R.N., Knick, V.B., Lackey, K., Lovejoy, B., Luzzio, M.J., Murray, D., Parker, P., Rocque, W.J., Shewchuk, L., Veal, J.M., Walker, D.H., Kuyper, L.F.(2001) Science 291: 134-137
- PubMed: 11141566
- DOI: https://doi.org/10.1126/science.291.5501.134
- Primary Citation of Related Structures:
1FVT, 1FVV - PubMed Abstract:
Most traditional cytotoxic anticancer agents ablate the rapidly dividing epithelium of the hair follicle and induce alopecia (hair loss). Inhibition of cyclin-dependent kinase 2 (CDK2), a positive regulator of eukaryotic cell cycle progression, may represent a therapeutic strategy for prevention of chemotherapy-induced alopecia (CIA) by arresting the cell cycle and reducing the sensitivity of the epithelium to many cell cycle-active antitumor agents. Potent small-molecule inhibitors of CDK2 were developed using structure-based methods. Topical application of these compounds in a neonatal rat model of CIA reduced hair loss at the site of application in 33 to 50% of the animals. Thus, inhibition of CDK2 represents a potentially useful approach for the prevention of CIA in cancer patients.
- Department of Cancer Biology, Glaxo Wellcome Research and Development, Research Triangle Park, NC 27709, USA. std41085@glaxowellcome.com
Organizational Affiliation: