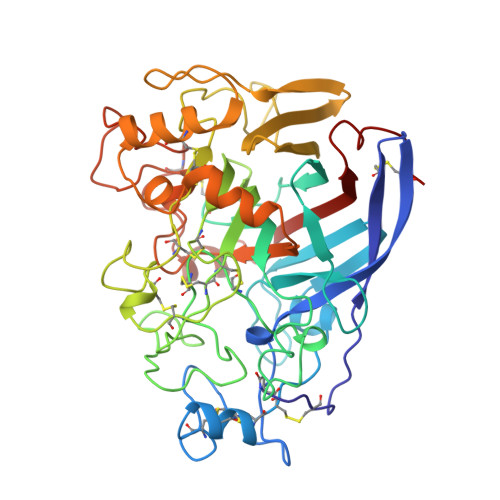
The three-dimensional crystal structure of the catalytic core of cellobiohydrolase I from Trichoderma reesei.
Divne, C., Stahlberg, J., Reinikainen, T., Ruohonen, L., Pettersson, G., Knowles, J.K., Teeri, T.T., Jones, T.A.(1994) Science 265: 524-528
- PubMed: 8036495
- DOI: https://doi.org/10.1126/science.8036495
- Primary Citation of Related Structures:
1CEL - PubMed Abstract:
Cellulose is the major polysaccharide of plants where it plays a predominantly structural role. A variety of highly specialized microorganisms have evolved to produce enzymes that either synergistically or in complexes can carry out the complete hydrolysis of cellulose. The structure of the major cellobiohydrolase, CBHI, of the potent cellulolytic fungus Trichoderma reesei has been determined and refined to 1.8 angstrom resolution. The molecule contains a 40 angstrom long active site tunnel that may account for many of the previously poorly understood macroscopic properties of the enzyme and its interaction with solid cellulose. The active site residues were identified by solving the structure of the enzyme complexed with an oligosaccharide, o-iodobenzyl-1-thio-beta-cellobioside. The three-dimensional structure is very similar to a family of bacterial beta-glucanases with the main-chain topology of the plant legume lectins.
- Department of Molecular Biology, Uppsala University, Sweden.
Organizational Affiliation: