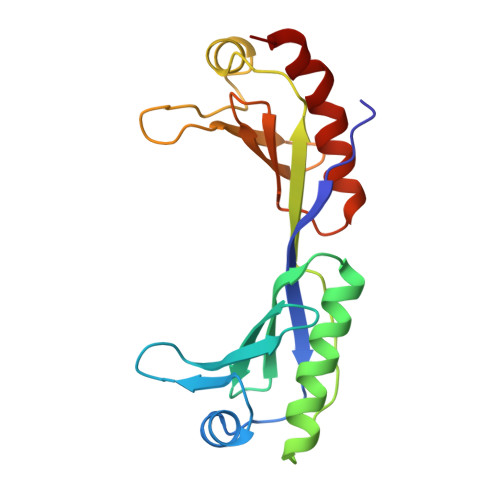
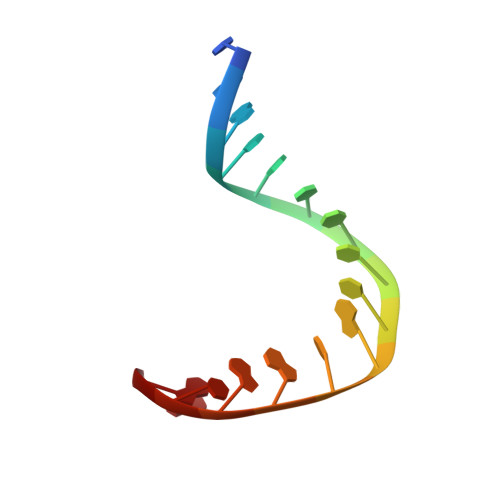
Crystal structure of a human TATA box-binding protein/TATA element complex.
Nikolov, D.B., Chen, H., Halay, E.D., Hoffman, A., Roeder, R.G., Burley, S.K.(1996) Proc Natl Acad Sci U S A 93: 4862-4867
- PubMed: 8643494
- DOI: https://doi.org/10.1073/pnas.93.10.4862
- Primary Citation of Related Structures:
1CDW - PubMed Abstract:
The TATA box-binding protein (TBP) is required by all three eukaryotic RNA polymerases for correct initiation of transcription of ribosomal, messenger, small nuclear, and transfer RNAs. The cocrystal structure of the C-terminal/core region of human TBP complexed with the TATA element of the adenovirus major late promoter has been determined at 1.9 angstroms resolution. Structural and functional analyses of the protein-DNA complex are presented, with a detailed comparison to our 1.9-angstroms resolution structure of Arabidopsis thaliana TBP2 bound to the same TATA box.
- Laboratory of Molecular Biophysics, The Rockefeller University, NY 10021, USA.
Organizational Affiliation: