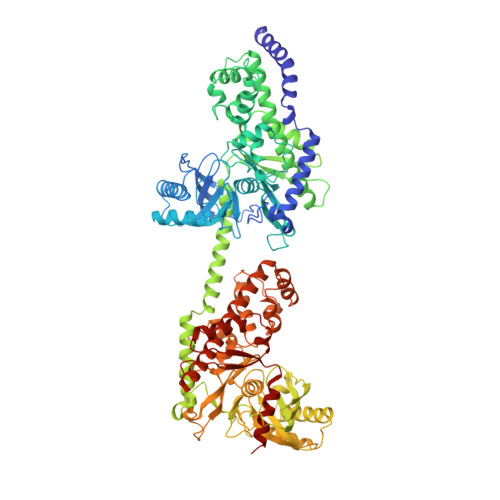
The structure of mammalian hexokinase-1.
Mulichak, A.M., Wilson, J.E., Padmanabhan, K., Garavito, R.M.(1998) Nat Struct Biol 5: 555-560
- PubMed: 9665168
- DOI: https://doi.org/10.1038/811
- Primary Citation of Related Structures:
1BDG, 1BG3 - PubMed Abstract:
We have determined the structures of the glucose-6-phosphate (G6P)-inhibitable 100,000 Mr Type I hexokinase from rat and the G6P-sensitive 50,000 Mr hexokinase from Schistosoma mansoni at a resolution of 2.8 and 2.6 A respectively. The structures define the glucose and G6P binding sites in these enzymes, suggest the mechanisms of intradomain G6P inhibition and activity loss in the Type I hexokinase N-terminal half, and reveal the structure of the membrane targeting motif that integrates the Type I hexokinase into the outer mitochondrial membrane.
- Department of Biochemistry, Michigan State University, East Lansing, USA. mulichak@alecto.bch.msu.edu
Organizational Affiliation: