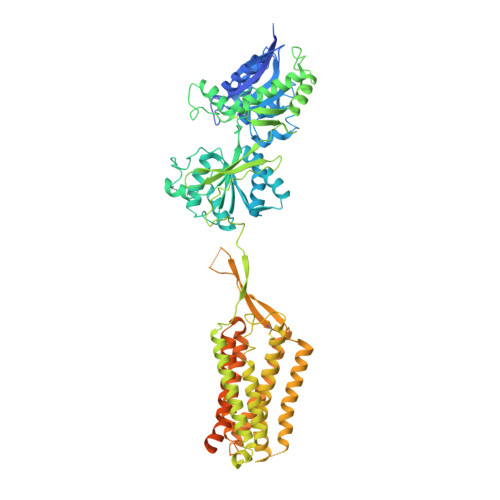
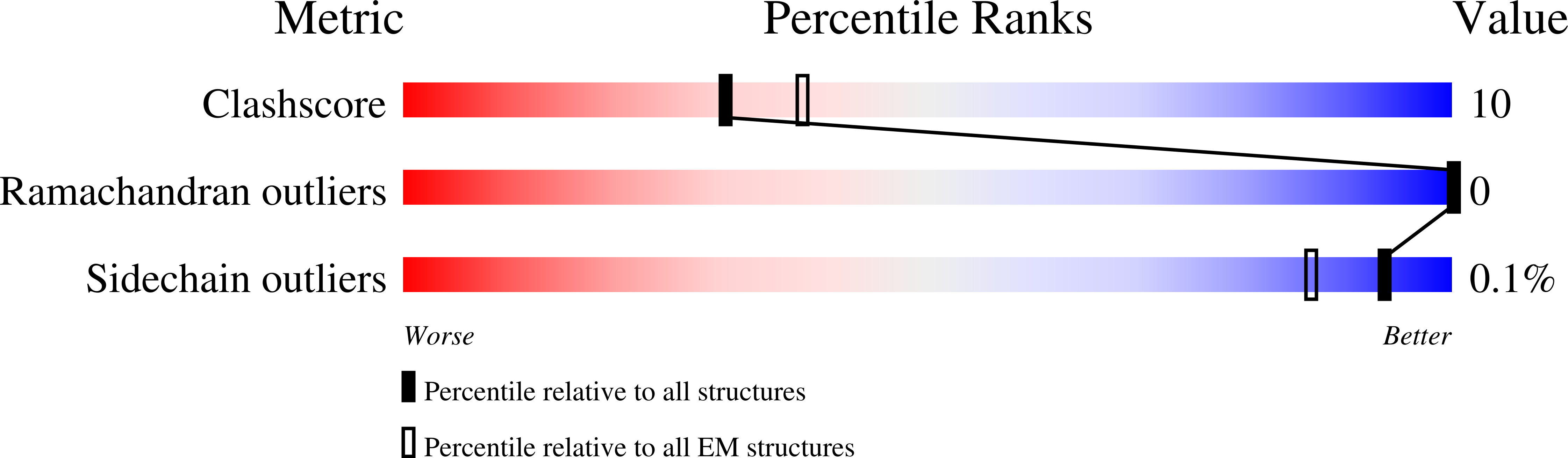Structures of metabotropic GABABreceptor.
Papasergi-Scott, M.M., Robertson, M.J., Seven, A.B., Panova, O., Mathiesen, J.M., Skiniotis, G.(2020) Nature 584: 310-314
- PubMed: 32580208
- DOI: https://doi.org/10.1038/s41586-020-2469-4
- Primary Citation of Related Structures:
6W2X, 6W2Y - PubMed Abstract:
Stimulation of the metabotropic GABA B receptor by γ-aminobutyric acid (GABA) results in prolonged inhibition of neurotransmission, which is central to brain physiology 1 . GABA B belongs to family C of the G-protein-coupled receptors, which operate as dimers to transform synaptic neurotransmitter signals into a cellular response through the binding and activation of heterotrimeric G proteins 2,3 . However, GABA B is unique in its function as an obligate heterodimer in which agonist binding and G-protein activation take place on distinct subunits 4,5 . Here we present cryo-electron microscopy structures of heterodimeric and homodimeric full-length GABA B receptors. Complemented by cellular signalling assays and atomistic simulations, these structures reveal that extracellular loop 2 (ECL2) of GABA B has an essential role in relaying structural transitions by ordering the linker that connects the extracellular ligand-binding domain to the transmembrane region. Furthermore, the ECL2 of each of the subunits of GABA B caps and interacts with the hydrophilic head of a phospholipid that occupies the extracellular half of the transmembrane domain, thereby providing a potentially crucial link between ligand binding and the receptor core that engages G proteins. These results provide a starting framework through which to decipher the mechanistic modes of signal transduction mediated by GABA B dimers, and have important implications for rational drug design that targets these receptors.
Organizational Affiliation:
Department of Molecular and Cellular Physiology, Stanford University School of Medicine, Stanford, CA, USA.