McsB forms a gated kinase chamber to mark aberrant bacterial proteins for degradation.
Hajdusits, B., Suskiewicz, M.J., Hundt, N., Meinhart, A., Kurzbauer, R., Leodolter, J., Kukura, P., Clausen, T.(2021) Elife 10
- PubMed: 34328418
- DOI: https://doi.org/10.7554/eLife.63505
- Primary Citation of Related Structures:
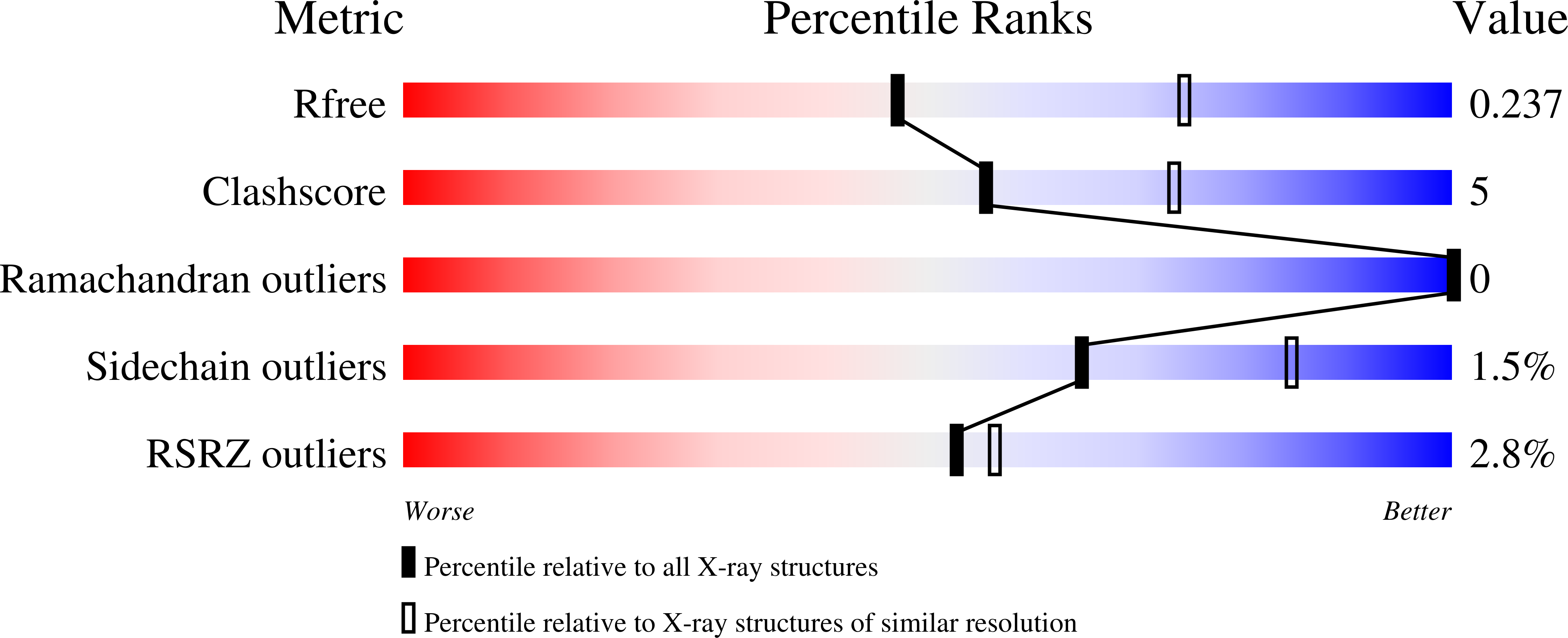
6TV6 - PubMed Abstract:
In Gram-positive bacteria, the McsB protein arginine kinase is central to protein quality control, labeling aberrant molecules for degradation by the ClpCP protease. Despite its importance for stress response and pathogenicity, it is still elusive how the bacterial degradation labeling is regulated. Here, we delineate the mechanism how McsB targets aberrant proteins during stress conditions. Structural data reveal a self-compartmentalized kinase, in which the active sites are sequestered in a molecular cage. The 'closed' octamer interconverts with other oligomers in a phosphorylation-dependent manner and, unlike these 'open' forms, preferentially labels unfolded proteins. In vivo data show that heat-shock triggers accumulation of higher order oligomers, of which the octameric McsB is essential for surviving stress situations. The interconversion of open and closed oligomers represents a distinct regulatory mechanism of a degradation labeler, allowing the McsB kinase to adapt its potentially dangerous enzyme function to the needs of the bacterial cell.
Organizational Affiliation:
Research Institute of Molecular Pathology (IMP), Vienna BioCenter, Vienna, Austria.