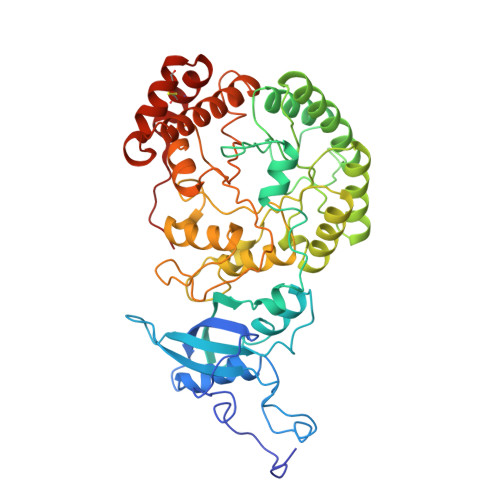
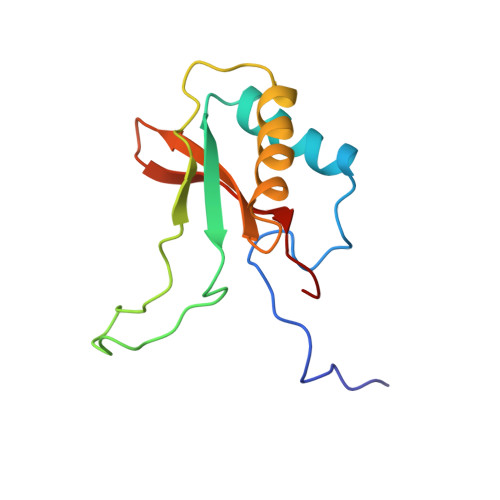
A crystal form of ribulose-1,5-bisphosphate carboxylase/oxygenase from Nicotiana tabacum in the activated state.
Suh, S.W., Cascio, D., Chapman, M.S., Eisenberg, D.(1987) J Mol Biol 197: 363-365
- PubMed: 3681999
- DOI: https://doi.org/10.1016/0022-2836(87)90129-x
- Primary Citation of Related Structures:
4RUB - PubMed Abstract:
A new crystal form of ribulose-1,5-bisphosphate carboxylase/oxygenase (EC 4.1.1.39) from Nicotiana tabacum has been obtained at alkaline pH with polyethylene glycol 8000 in the presence of a non-ionic detergent, beta-octyl glucoside. The crystals are grown at room temperature by the hanging-drop vapor diffusion technique from a protein solution containing enzyme complexed with CO2, Mg2+, and the transition state analog 2-C-carboxy-D-arabinitol-1,5-bisphosphate. The crystals belong to the the space group P3(1)21 (or P3(2)21) with the cell parameters a = 204.6 A, and c = 117.4 A (1 A = 0.1 nm). The asymmetric unit contains half (L4S4: L, large subunit, 53,000 Mr; S, small subunit, 15,000 Mr) of a hexadecameric molecule (L8S8, 540,000 Mr). The crystals diffract to at least 2.6 A Bragg spacing and are suitable for X-ray structure determination.
Organizational Affiliation:
Molecular Biology Institute, University of California, Los Angeles 90024.