Structural and Dynamic Characterization of a Vinculin Binding Site in the Talin Rod
Gingras, A.R., Vogel, K.P., Steinhoff, H.J., Ziegler, W.H., Patel, B., Emsley, J., Critchley, D.R., Roberts, G.C., Barsukov, I.L.(2006) Biochemistry 45: 1805-1817
- PubMed: 16460027
- DOI: https://doi.org/10.1021/bi052136l
- Primary Citation of Related Structures:
1XWJ, 2B0H - PubMed Abstract:
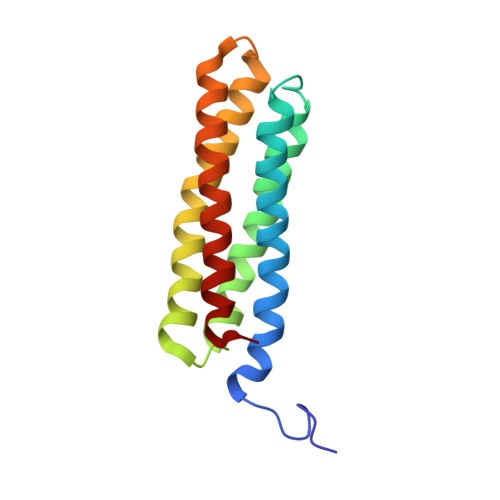
Talin is a key protein involved in linking integrins to the actin cytoskeleton. The long flexible talin rod domain contains a number of binding sites for vinculin, a cytoskeletal protein important in stabilizing integrin-mediated cell-matrix junctions. Here we report the solution structure of a talin rod polypeptide (residues 1843-1973) which contains a single vinculin binding site (VBS; residues 1944-1969). Like other talin rod polypeptides, it consists of a helical bundle, in this case a four-helix bundle with a right-handed topology. The residues in the VBS important for vinculin binding were identified by studying the binding of a series of VBS-related peptides to the vinculin Vd1 domain. The key binding determinants are buried in the interior of the helical bundle, suggesting that a substantial structural change in the talin polypeptide is required for vinculin binding. Direct evidence for this was obtained by NMR and EPR spectroscopy. [1H,15N]-HSQC spectra of the talin fragment indicate that vinculin binding caused approximately two-thirds of the protein to adopt a flexible random coil. For EPR spectroscopy, nitroxide spin labels were attached to the talin polypeptide via appropriately located cysteine residues. Measurements of inter-nitroxide distances in doubly spin-labeled protein showed clearly that the helical bundle is disrupted and the mobility of the helices, except for the VBS helix, is markedly increased. Binding of vinculin to talin is thus a clear example of the unusual phenomenon of protein unfolding being required for protein/protein interaction.
Organizational Affiliation:
Department of Biochemistry, University of Leicester, Henry Wellcome Building, P.O. Box 138, Lancaster Road, Leicester LE1 9HN, United Kingdom.