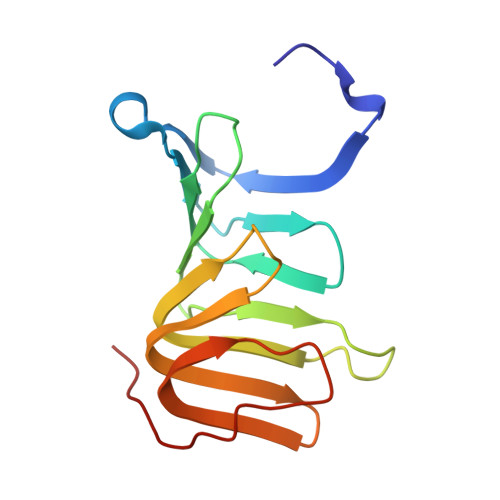
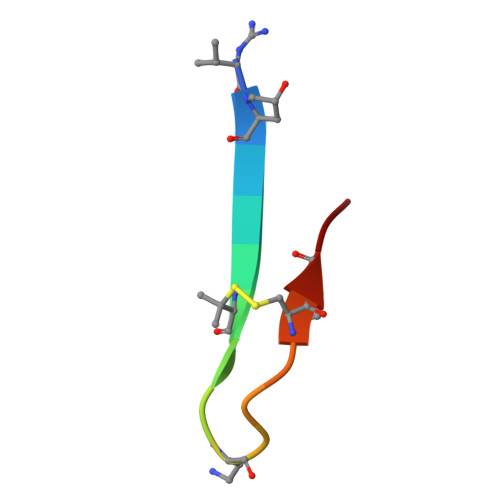
Early Molecular Insights into Thanatin Analogues Binding to A. baumannii LptA.
Oi, K.K., Moehle, K., Schuster, M., Zerbe, O.(2023) Molecules 28
- PubMed: 37298811
- DOI: https://doi.org/10.3390/molecules28114335
- Primary Citation of Related Structures:
8ONU - PubMed Abstract:
The cationic antimicrobial ß-hairpin, thanatin, was recently developed into drug-like analogues active against carbapenem-resistant Enterobacteriaceae (CRE). The analogues represent new antibiotics with a novel mode of action targeting LptA in the periplasm and disrupting LPS transport. The compounds lose antimicrobial efficacy when the sequence identity to E. coli LptA falls below 70%. We wanted to test the thanatin analogues against LptA of a phylogenetic distant organism and investigate the molecular determinants of inactivity. Acinetobacter baumannii ( A. baumannii ) is a critical Gram-negative pathogen that has gained increasing attention for its multi-drug resistance and hospital burden. A. baumannii LptA shares 28% sequence identity with E. coli LptA and displays an intrinsic resistance to thanatin and thanatin analogues (MIC values > 32 µg/mL) through a mechanism not yet described. We investigated the inactivity further and discovered that these CRE-optimized derivatives can bind to LptA of A. baumannii in vitro, despite the high MIC values. Herein, we present a high-resolution structure of A. baumannii LptAm in complex with a thanatin derivative 7 and binding affinities of selected thanatin derivatives. Together, these data offer structural insights into why thanatin derivatives are inactive against A. baumannii LptA, despite binding events in vitro.
Organizational Affiliation:
Department of Chemistry, University of Zurich, Winterthurerstrasse 190, CH-8057 Zurich, Switzerland.