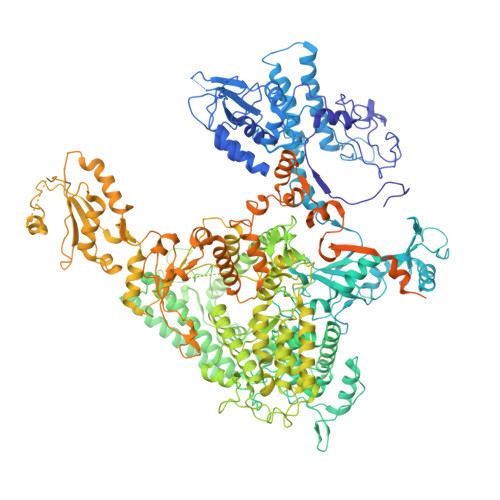
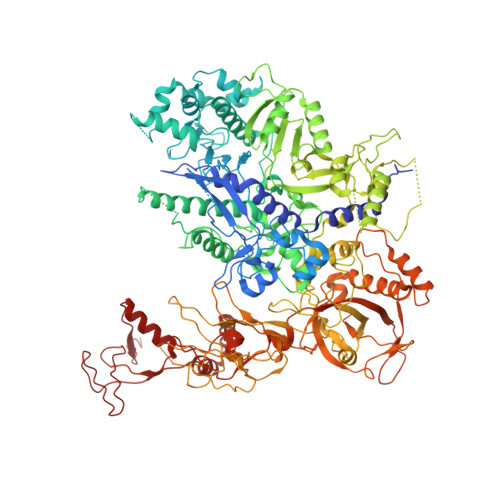
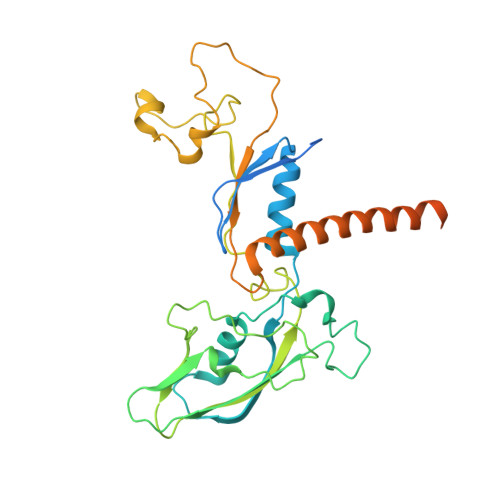
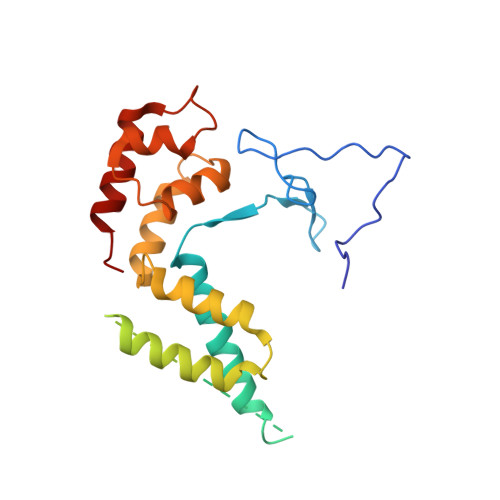
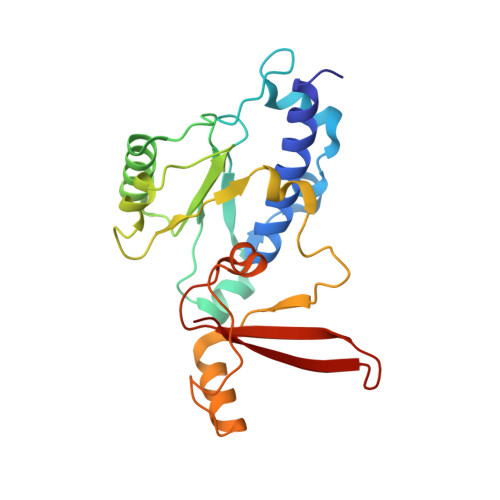
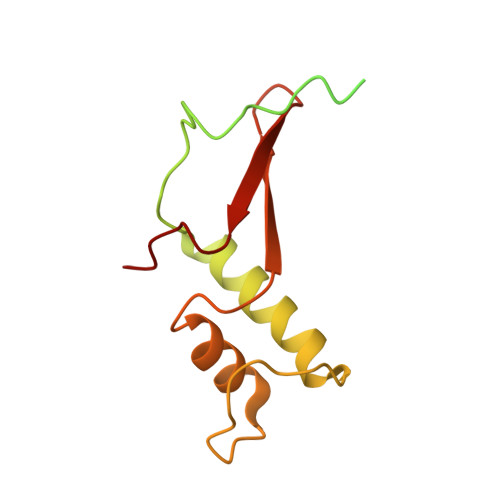
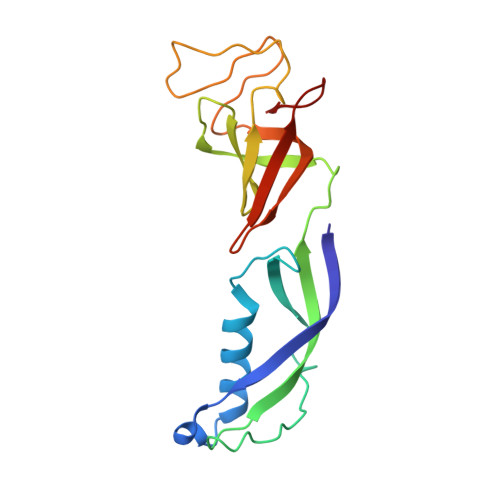
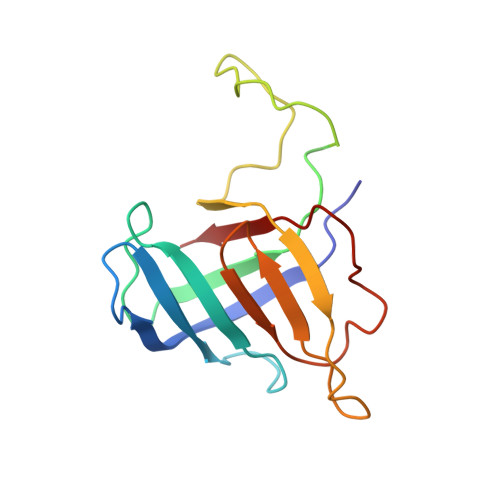
Structure of transcribing RNA polymerase II-nucleosome complex.
Farnung, L., Vos, S.M., Cramer, P.(2018) Nat Commun 9: 5432-5432
- PubMed: 30575770
- DOI: https://doi.org/10.1038/s41467-018-07870-y
- Primary Citation of Related Structures:
6I84 - PubMed Abstract:
Transcription of eukaryotic protein-coding genes requires passage of RNA polymerase II (Pol II) through nucleosomes, but it is unclear how this is achieved. Here we report the cryo-EM structure of transcribing Saccharomyces cerevisiae Pol II engaged with a downstream nucleosome core particle at an overall resolution of 4.4 Å. Pol II and the nucleosome are observed in a defined relative orientation that is not predicted. Pol II contacts both sides of the nucleosome dyad using its clamp head and lobe domains. Structural comparisons reveal that the elongation factors TFIIS, DSIF, NELF, SPT6, and PAF1 complex can be accommodated on the Pol II surface in the presence of the oriented nucleosome. Our results provide a starting point for analysing the mechanisms of chromatin transcription.
Organizational Affiliation:
Max Planck Institute for Biophysical Chemistry, Department of Molecular Biology, Am Fassberg 11, 37077, Göttingen, Germany.