Structural Evidence for a Role of the Multi-functional Human Glycoprotein Afamin in Wnt Transport.
Naschberger, A., Orry, A., Lechner, S., Bowler, M.W., Nurizzo, D., Novokmet, M., Keller, M.A., Oemer, G., Seppi, D., Haslbeck, M., Pansi, K., Dieplinger, H., Rupp, B.(2017) Structure 25: 1907-1915.e5
- PubMed: 29153507
- DOI: https://doi.org/10.1016/j.str.2017.10.006
- Primary Citation of Related Structures:
5OKL - PubMed Abstract:
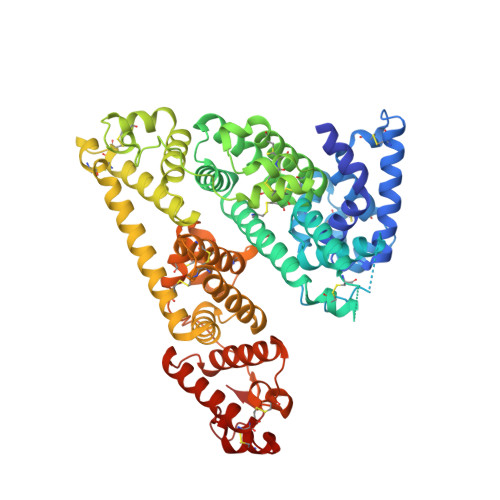
Afamin, a human plasma glycoprotein and putative transporter of hydrophobic molecules, has been shown to act as extracellular chaperone for poorly soluble, acylated Wnt proteins, forming a stable, soluble complex with functioning Wnt proteins. The 2.1-Å crystal structure of glycosylated human afamin reveals an almost exclusively hydrophobic binding cleft capable of harboring large hydrophobic moieties. Lipid analysis confirms the presence of lipids, and density in the primary binding pocket of afamin was modeled as palmitoleic acid, presenting the native O-acylation on serine 209 in human Wnt3a. The modeled complex between the experimental afamin structure and a Wnt3a homology model based on the XWnt8-Fz8-CRD fragment complex crystal structure is compelling, with favorable interactions comparable with the crystal structure complex. Afamin readily accommodates the conserved palmitoylated serine 209 of Wnt3a, providing a structural basis how afamin solubilizes hydrophobic and poorly soluble Wnt proteins.
Organizational Affiliation:
Division of Genetic Epidemiology, Medical University of Innsbruck, Schöpfstraße 41, 6020 Innsbruck, Austria; Division of Biological Chemistry, Medical University of Innsbruck, Innrain 80-82, 6020 Innsbruck, Austria.