Structure, activity and thermostability investigations of OXA-163, OXA-181 and OXA-245 using biochemical analysis, crystal structures and differential scanning calorimetry analysis.
Lund, B.A., Thomassen, A.M., Carlsen, T.J.O., Leiros, H.K.S.(2017) Acta Crystallogr F Struct Biol Commun 73: 579-587
- PubMed: 28994407
- DOI: https://doi.org/10.1107/S2053230X17013838
- Primary Citation of Related Structures:
5ODZ, 5OE0, 5OE2 - PubMed Abstract:
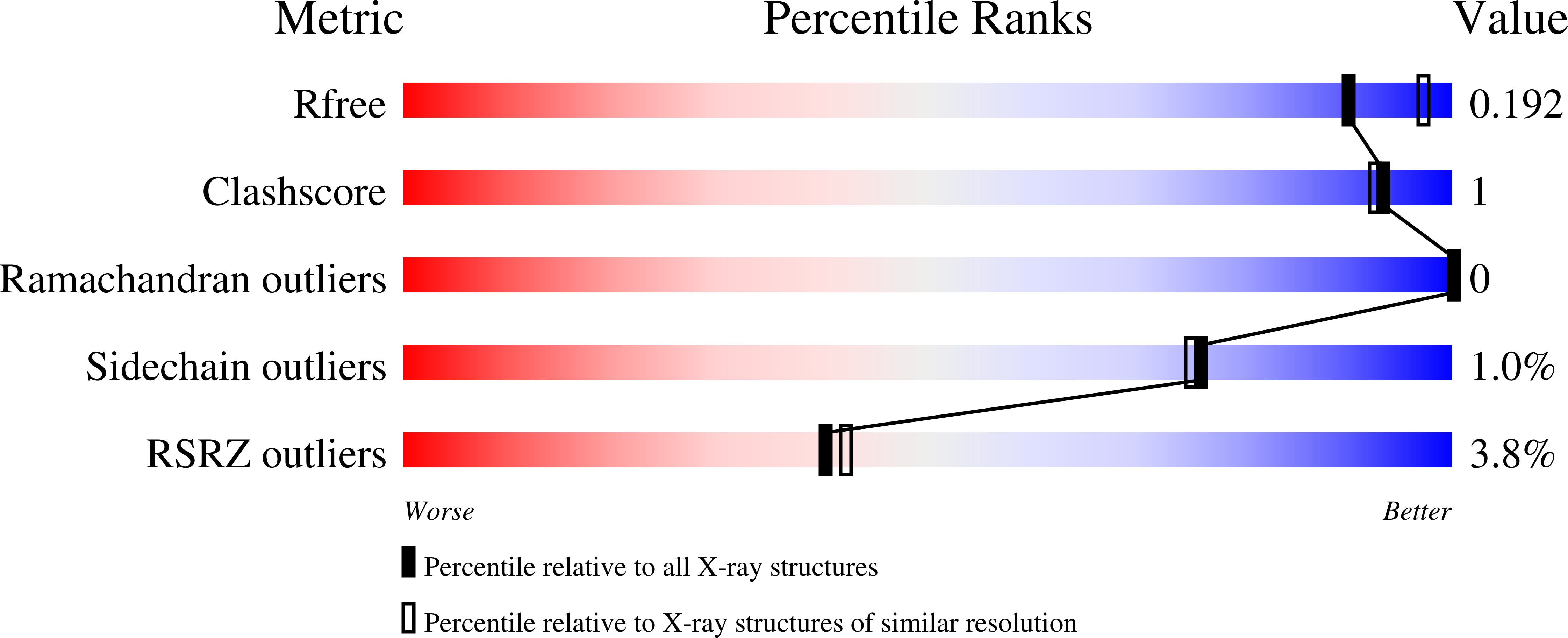
The first crystal structures of the class D β-lactamases OXA-181 and OXA-245 were determined to 2.05 and 2.20 Å resolution, respectively; in addition, the structure of a new crystal form of OXA-163 was resolved to 2.07 Å resolution. All of these enzymes are OXA-48-like and have been isolated from different clinical Klebsiella pneumoniae strains and also from other human pathogens such as Pseudomonas aeruginosa and Escherichia coli. Here, enzyme kinetics and thermostability studies are presented, and the new crystal structures are used to explain the observed variations. OXA-245 had the highest melting point (T m = 55.8°C), as determined by differential scanning calorimetry, compared with OXA-163 (T m = 49.4°C) and OXA-181 (T m = 52.6°C). The differences could be explained by the loss of two salt bridges in OXA-163, and an overall decrease in the polarity of the surface of OXA-181 compared with OXA-245.
Organizational Affiliation:
Department of Chemistry, UiT The Arctic University of Norway, 9037 Tromsø, Norway.