The Crystal Structure of the C-Terminal Domain of the Salmonella enterica PduO Protein: An Old Fold with a New Heme-Binding Mode.
Ortiz de Orue Lucana, D., Hickey, N., Hensel, M., Klare, J.P., Geremia, S., Tiufiakova, T., Torda, A.E.(2016) Front Microbiol 7: 1010-1010
- PubMed: 27446048
- DOI: https://doi.org/10.3389/fmicb.2016.01010
- Primary Citation of Related Structures:
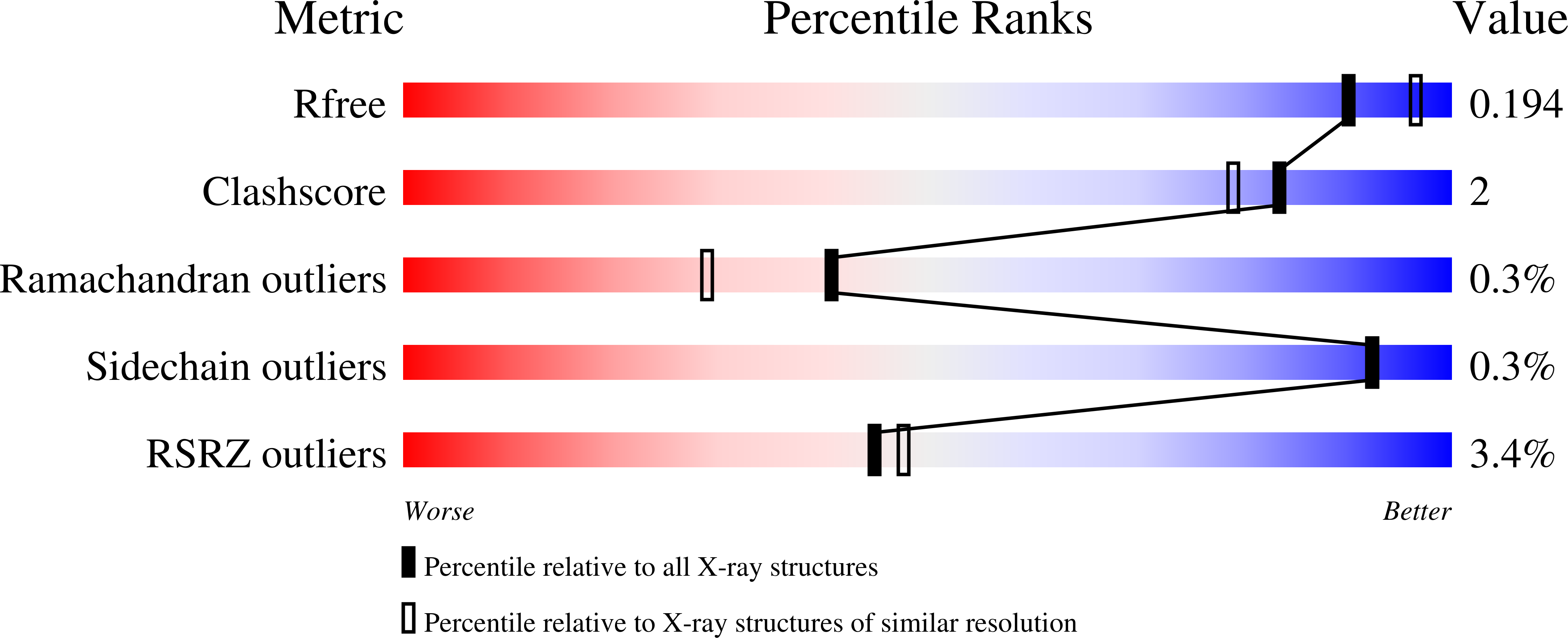
5CX7 - PubMed Abstract:
The two-domain protein PduO, involved in 1,2-propanediol utilization in the pathogenic Gram-negative bacterium Salmonella enterica is an ATP:Cob(I)alamin adenosyltransferase, but this is a function of the N-terminal domain alone. The role of its C-terminal domain (PduOC) is, however, unknown. In this study, comparative growth assays with a set of Salmonella mutant strains showed that this domain is necessary for effective in vivo catabolism of 1,2-propanediol. It was also shown that isolated, recombinantly-expressed PduOC binds heme in vivo. The structure of PduOC co-crystallized with heme was solved (1.9 Å resolution) showing an octameric assembly with four heme moieities. The four heme groups are highly solvent-exposed and the heme iron is hexa-coordinated with bis-His ligation by histidines from different monomers. Static light scattering confirmed the octameric assembly in solution, but a mutation of the heme-coordinating histidine caused dissociation into dimers. Isothermal titration calorimetry using the PduOC apoprotein showed strong heme binding (K d = 1.6 × 10(-7) M). Biochemical experiments showed that the absence of the C-terminal domain in PduO did not affect adenosyltransferase activity in vitro. The evidence suggests that PduOC:heme plays an important role in the set of cobalamin transformations required for effective catabolism of 1,2-propanediol. Salmonella PduO is one of the rare proteins which binds the redox-active metabolites heme and cobalamin, and the heme-binding mode of the C-terminal domain differs from that in other members of this protein family.
Organizational Affiliation:
Applied Genetics of Microorganisms, Department of Biology/Chemistry, University of Osnabrück Osnabrück, Germany.