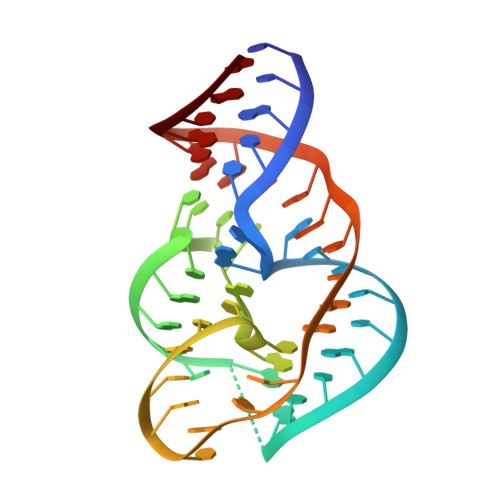
Crystal structure and mechanistic investigation of the twister ribozyme.
Liu, Y., Wilson, T.J., McPhee, S.A., Lilley, D.M.(2014) Nat Chem Biol 10: 739-744
- PubMed: 25038788
- DOI: https://doi.org/10.1038/nchembio.1587
- Primary Citation of Related Structures:
4OJI - PubMed Abstract:
We present a crystal structure at 2.3-Å resolution of the recently described nucleolytic ribozyme twister. The RNA adopts a previously uncharacterized compact fold based on a double-pseudoknot structure, with the active site at its center. Eight highly conserved nucleobases stabilize the core of the ribozyme through the formation of one Watson-Crick and three noncanonical base pairs, and the highly conserved adenine 3' of the scissile phosphate is bound in the major groove of an adjacent pseudoknot. A strongly conserved guanine nucleobase directs its Watson-Crick edge toward the scissile phosphate in the crystal structure, and mechanistic evidence supports a role for this guanine as either a general base or acid in a concerted, general acid-base-catalyzed cleavage reaction.
Organizational Affiliation:
Cancer Research UK Nucleic Acid Structure Research Group, Medical Sciences Institute/Wellcome Trust Building (MSI/WTB) Complex, University of Dundee, Dundee, UK.