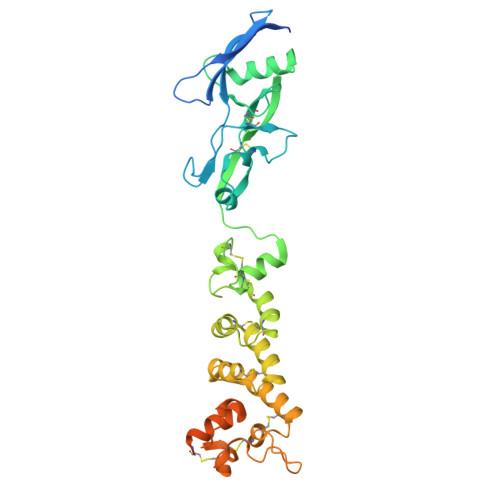
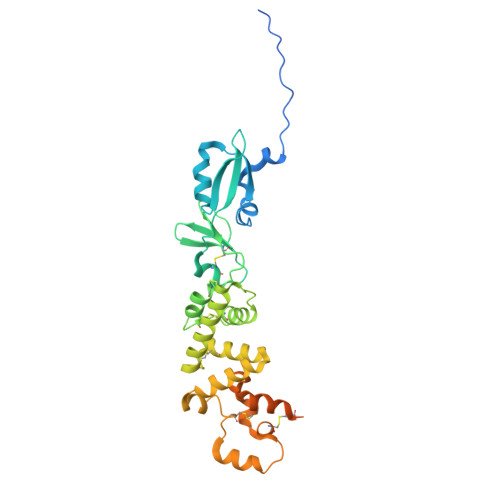
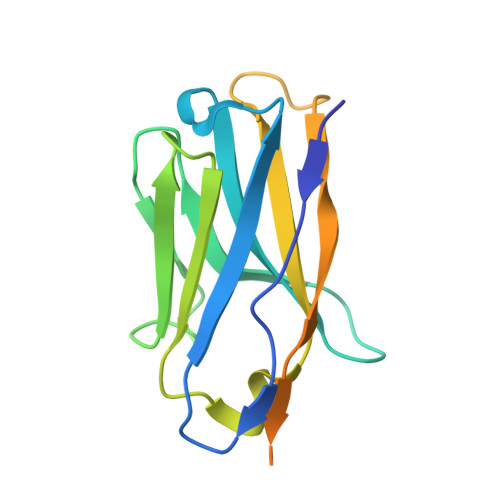
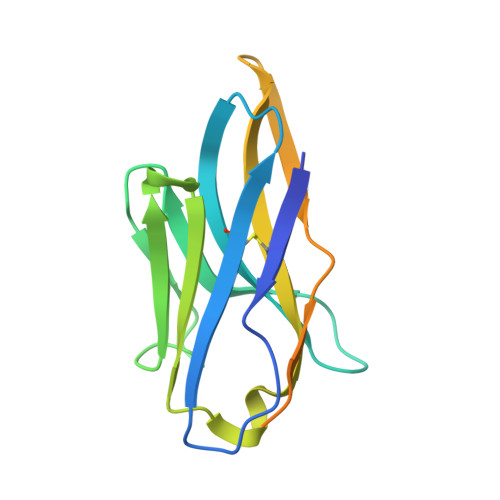
Structural basis of poxvirus fusion regulation and anti-A16/G9 antibody-mediated neutralization and protection.
Meola, A., Vernuccio, R., Battini, L., Albericio, G., Delgado, P., Bamford, R., Pokorny, L., Broutin, M., Martinez Leon, A., Gallien, S., Gil, M., Noriega, M.A., Guivel-Benhassine, F., Porrot, F., Postal, J., Buchrieser, J., Hubert, M., Haouz, A., Lafaye, P., Esteban, M., Hub, J.S., Mahevas, M., Chappert, P., Mercer, J., Garcia-Arriaza, J., Schwartz, O., Guardado-Calvo, P.(2025) Cell 188: 6266
- PubMed: 40865523
- DOI: https://doi.org/10.1016/j.cell.2025.07.040
- Primary Citation of Related Structures:
9HBK, 9HL2, 9HLS, 9HNG, 9HPA, 9R09, 9R0B, 9R0J, 9RDH - PubMed Abstract:
Monkeypox virus (MPXV) is a poxvirus endemic to Central and West Africa with high epidemic potential. Poxviruses enter host cells via a conserved entry-fusion complex (EFC), which mediates viral fusion to the cell membrane. The EFC is a promising therapeutic target, but the absence of structural data has limited the development of fusion-inhibiting treatments. Here, we investigated A16/G9, a subcomplex of the EFC that controls fusion timing. Using cryo-electron microscopy, we showed how A16/G9 interacts with A56/K2, a viral fusion suppressor that prevents superinfection. Immunization with A16/G9 elicited a protective immune response in mice. Using X-ray crystallography, we characterized two neutralizing antibodies and engineered a chimeric antibody that cross-neutralizes several poxviruses more efficiently than 7D11, the most potent antibody targeting the EFC described to date. These findings highlight the potential of A16/G9 as a candidate for subunit vaccines and identify regions of the EFC as targets for antiviral development.
- Structural biology of infectious diseases G5+ unit, Institut Pasteur, Université Paris Cité, Paris, France.
Organizational Affiliation: