Cryo-EM reconstruction of PI3KC3-C2 in complex with Rubicon Middle Region of C terminus
Chen, M., Ni, Q., Hurley, J.H.To be published.
Experimental Data Snapshot
Starting Model: in silico
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Phosphoinositide 3-kinase regulatory subunit 4 | 1,445 | Homo sapiens | Mutation(s): 0 Gene Names: PIK3R4 EC: 2.7.11.1 | 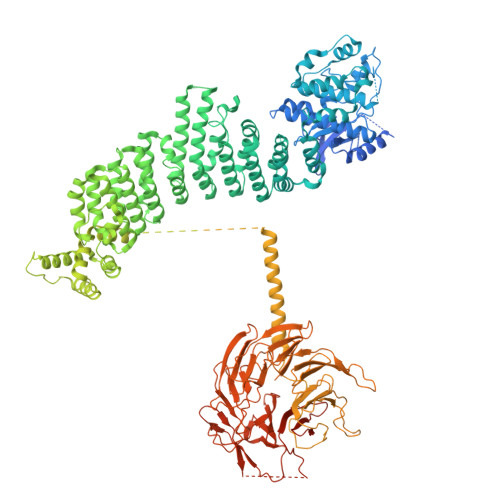 | |
UniProt | |||||
Find proteins for A0JP11 (Homo sapiens) Explore A0JP11 Go to UniProtKB: A0JP11 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0JP11 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Phosphatidylinositol 3-kinase catalytic subunit type 3 | 887 | Homo sapiens | Mutation(s): 0 Gene Names: PIK3C3, VPS34 EC: 2.7.1.137 | 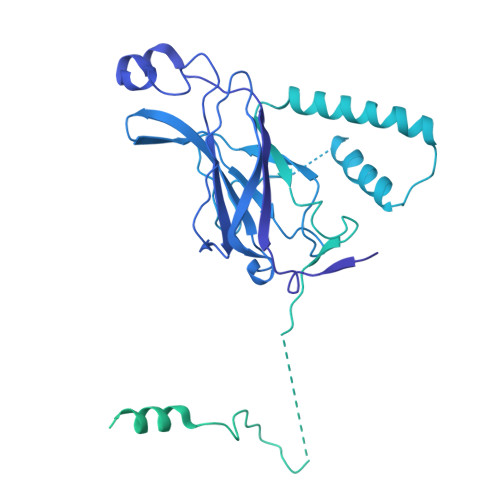 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q8NEB9 (Homo sapiens) Explore Q8NEB9 Go to UniProtKB: Q8NEB9 | |||||
PHAROS: Q8NEB9 GTEx: ENSG00000078142 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8NEB9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| UV radiation resistance-associated gene protein | 699 | Homo sapiens | Mutation(s): 0 Gene Names: UVRAG | 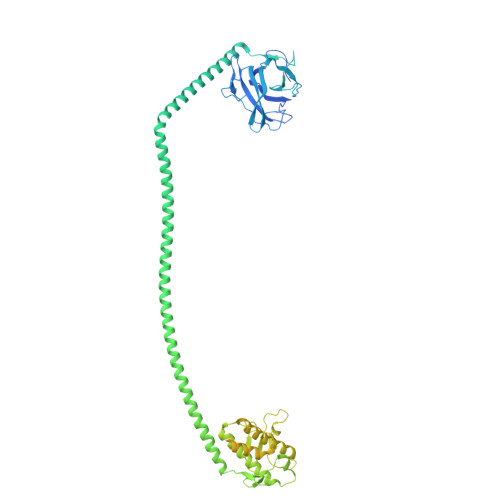 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9P2Y5 (Homo sapiens) Explore Q9P2Y5 Go to UniProtKB: Q9P2Y5 | |||||
PHAROS: Q9P2Y5 GTEx: ENSG00000198382 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9P2Y5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Beclin-1,Run domain Beclin-1-interacting and cysteine-rich domain-containing protein chimera | 663 | Homo sapiens | Mutation(s): 0 Gene Names: BECN1, GT197, RUBCN, KIAA0226 | 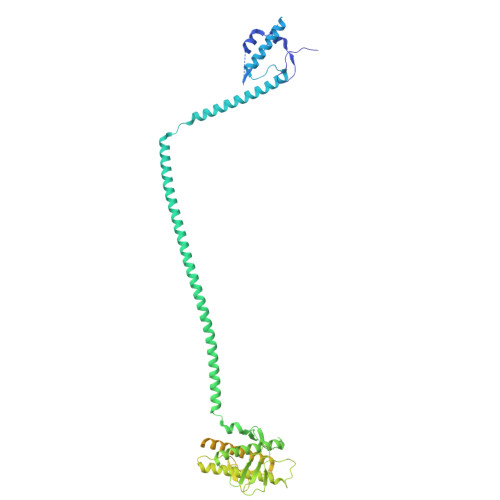 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q14457 (Homo sapiens) Explore Q14457 Go to UniProtKB: Q14457 | |||||
PHAROS: Q14457 GTEx: ENSG00000126581 | |||||
Find proteins for Q92622 (Homo sapiens) Explore Q92622 Go to UniProtKB: Q92622 | |||||
PHAROS: Q92622 GTEx: ENSG00000145016 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Groups | Q14457Q92622 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GTP (Subject of Investigation/LOI) Query on GTP | F [auth A] | GUANOSINE-5'-TRIPHOSPHATE C10 H16 N5 O14 P3 XKMLYUALXHKNFT-UUOKFMHZSA-N |  | ||
| ZN (Subject of Investigation/LOI) Query on ZN | G [auth D] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.21.2_5419 |
| RECONSTRUCTION | cryoSPARC | 4.5.3 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of Neurological Disorders and Stroke (NIH/NINDS) | United States | NS134598 |
| Michael J. Fox Foundation | United States | ASAP-000350 |