Structural Insights into the Glycosylphosphatidylinositol Mannosyltransferase I Complex from Candida glabrata .
Sun, H., Wu, W., Li, X., Deng, Y., Huang, J., Yin, M., Yan, Z.(2025) J Fungi (Basel) 11
- PubMed: 41295199
- DOI: https://doi.org/10.3390/jof11110819
- Primary Citation of Related Structures:
9WNO - PubMed Abstract:
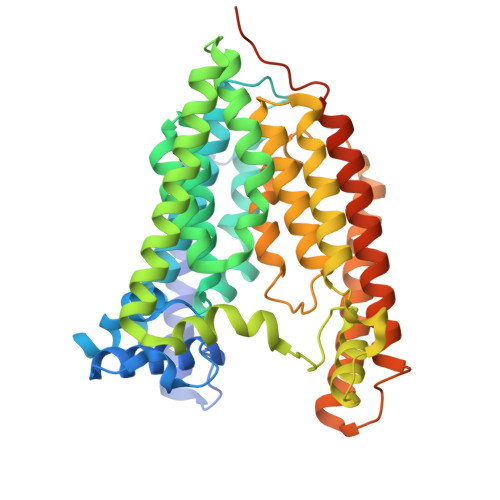
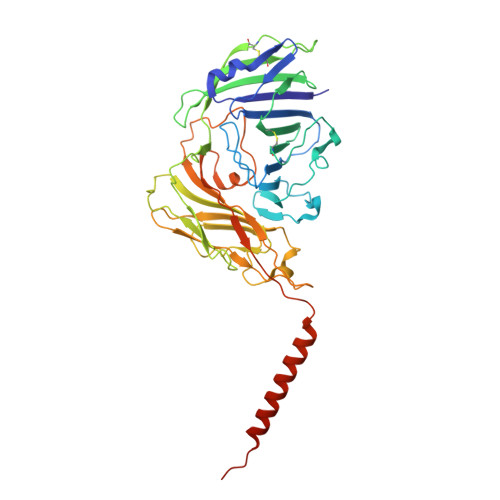
The global rise in resistance to first-line antifungal agents highlights the urgent need for new therapeutic strategies. Glycosylphosphatidylinositol (GPI)-anchored protein biosynthesis is an attractive target. The GPI mannosyltransferase I (GPI-MT-I), composed of Gpi14 and Pbn1, catalyzes the essential first mannose transfer from dolichol-phosphomannose (Dol-P-Man) to the GPI precursor. This initial mannosylation is critical for fungal cell wall integrity, yet the molecular basis of GPI-MT-I assembly and substrate recognition remains poorly understood. Here, we present the cryo-EM structure of Candida glabrata GPI-MT-I in complex with Dol-P-Man, revealing how Gpi14 and Pbn1 form a stable complex and engage the mannose donor. An AlphaFold3-predicted acceptor-bound model further defines the structural basis of acceptor substrate recognition and suggests a plausible catalytic mechanism. Comparison with structural homologs highlights a distinct mode of substrate engagement by GPI-MT-I. Together, these findings establish a mechanistic framework for GPI-MT-I function with broader implications for the GPI-MT family.
- School of Biomedical Sciences, Hunan University, Changsha 410082, China.
Organizational Affiliation: