Discovery of ART5537: A Potent and Selective Small-Molecule Probe for EXO1.
Mann, S.E., Davis, O.A., Bomke, J., Cornaciu, I., Elinati, E., Follows, B., Galbiati, A., Geo, L., Grande, D., Jorand-Lebrun, C., Lademann, C.A., Lefranc, J., Leuthner, B., Mason, B., McWhirter, C.L., Musil, D., Pehl, U., Petersson, C., Pica, A., Pinto, M.F., Rajendra, E., Rakers, C., Rego, A.T., Robinson, H.M.R., Schwarz, D., Smith, G.C.M., Sorrell, F.J., Zenke, F.T., Heald, R.A., Burgdorf, L.T.(2025) J Med Chem 68: 26432-26447
- PubMed: 41359073
- DOI: https://doi.org/10.1021/acs.jmedchem.5c02593
- Primary Citation of Related Structures:
9SEB, 9SMO - PubMed Abstract:
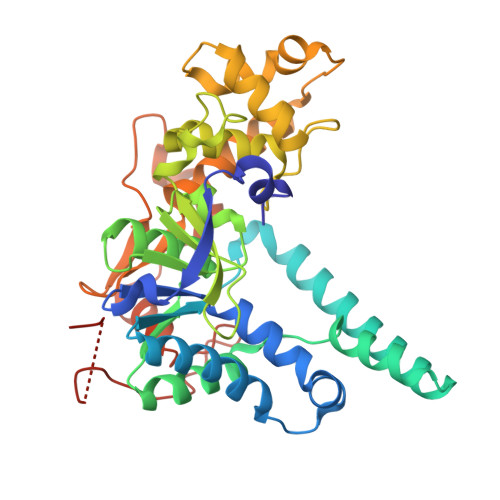
Exonuclease 1 (EXO1) is emerging as a target of interest in oncology due to its involvement in multifaceted DNA metabolic processes, particularly in homologous recombination (HR). Evidence is building that BRCA1 -deficient cancers are sensitive to loss of EXO1, suggesting therapeutic potential for treating certain subsets of patients. However, EXO1 remains under-explored, with very few reported inhibitors, and there is a paucity of good quality, potent, and selective pharmacological tools to explore its biology. Here, we describe a metal-chelating fragment screen, which resulted in highly selective, submicromolar EXO1 hits. Our subsequent structure-based design and optimization led to the discovery of ART5537 , the first highly potent and selective EXO1 inhibitor. We demonstrate that inhibition of EXO1 leads to potent suppression of HR in cells and that the HR inhibition of ART5537 is driven exclusively by EXO1. Furthermore, we show that ART5537 sensitizes cancer cells to ionizing radiation (IR) and synergizes with PARP inhibitors (PARPi).
- Artios Pharma Ltd., B940, Babraham Research Campus, Cambridge CB22 3FH, U.K.
Organizational Affiliation: