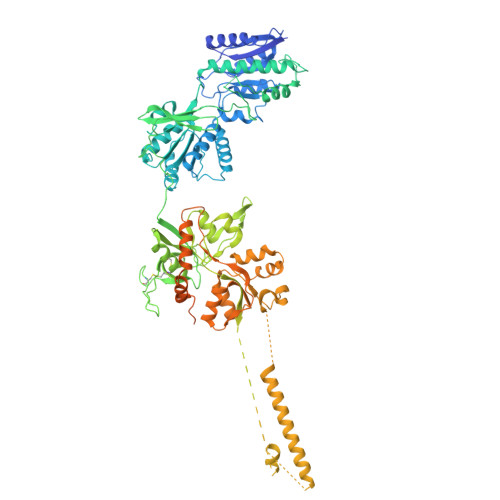
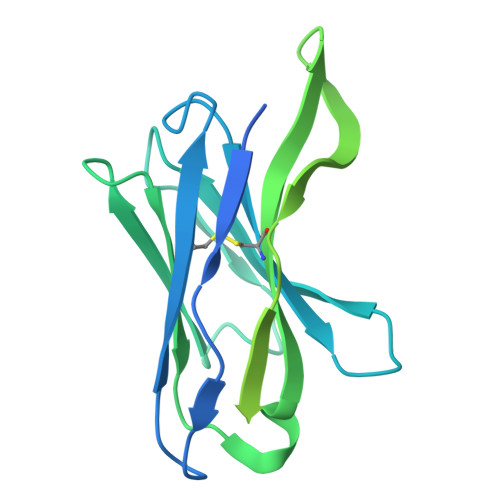
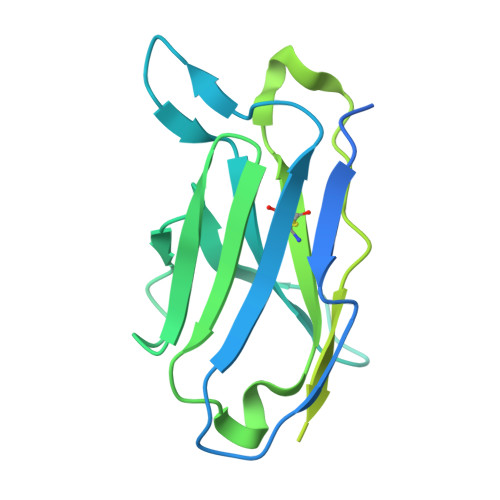
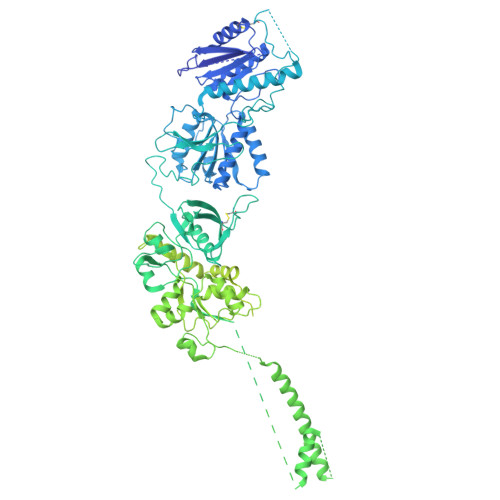
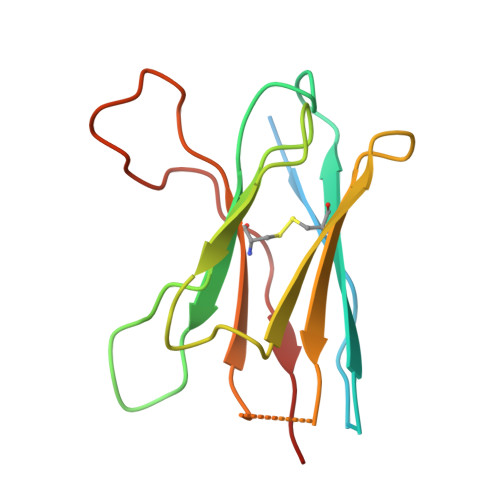
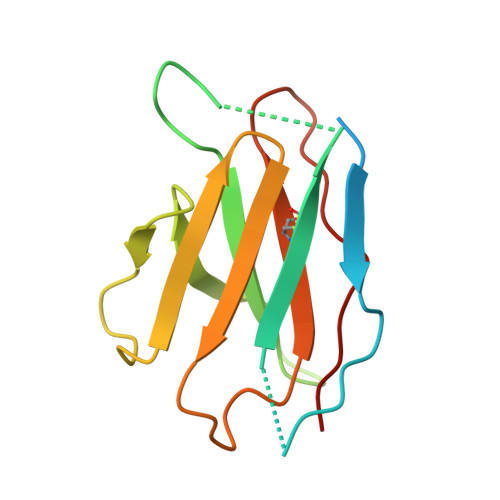
Cryo-EM of autoantibody-bound NMDA receptors reveals antigenic hotspots in an active immunization model of anti-NMDAR encephalitis.
Kim, J., Jalali-Yazdi, F., Jones, B.E., Westbrook, G.L., Gouaux, E.(2026) Sci Adv 12: eaeb4249-eaeb4249
- PubMed: 41533802
- DOI: https://doi.org/10.1126/sciadv.aeb4249
- Primary Citation of Related Structures:
9PZQ, 9PZR, 9PZS, 9PZT, 9PZU, 9PZV, 9PZW, 9PZX - PubMed Abstract:
Autoantibodies targeting synaptic membrane proteins are associated with autoimmune encephalitis manifested by seizures, psychosis, and memory dysfunction. Anti- N -methyl-d-aspartate receptor (NMDAR) encephalitis, a prototype of these autoimmune synaptic disorders, is unexpectedly common. Unfortunately, how the native repertoire of anti-NMDAR autoantibodies recognizes NMDARs and the precise locations of antigenic epitopes remain poorly understood. Here, we used an active immunization model that closely mimics the human disease to immunize adult mice with intact GluN1/GluN2A receptors, resulting in fulminant autoimmune encephalitis. Serum was collected at 6 weeks postimmunization for single-particle cryo-electron microscopy of GluN1/GluN2A receptors complexed with purified polyclonal anti-NMDAR autoantibody fragments. Native autoantibodies recognized two distinct binding sites on the GluN1 amino-terminal domain, which we confirmed using monoclonal antibodies bound to native NMDARs purified from mouse brain. Structural analysis of autoantibody-bound NMDAR complexes identified antigenic hotspots within the GluN1 amino-terminal domain. These hotspots provide potential targets for therapeutic intervention.
- Vollum Institute, Oregon Health & Science University, 3232 SW Research Drive, Portland, OR 97239, USA.
Organizational Affiliation: