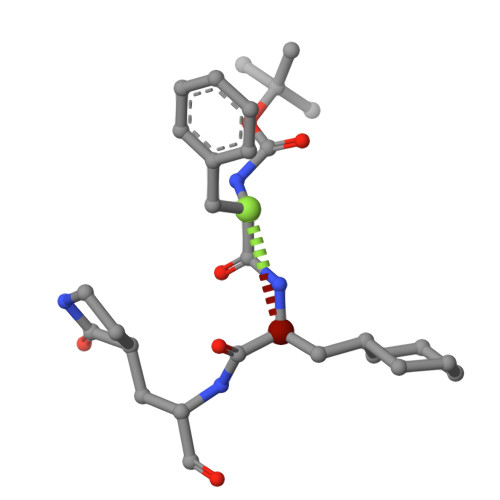
Broad-Spectrum Peptidomimetic Inhibitors of Norovirus and Coronavirus 3C-like Proteases.
Brimble, M.A., Stubbing, L.A., Hermant, Y.O., Yang, S.H., Hubert, J.G., Pearl, E.S., McSweeney, A.M., Young, V.L., Campbell, A.C., Opel-Reading, H.K., McKenzie-Goldsmith, G.M., Putha, L., Mortuza, R., Wang, H., Hurst, B.L., Julander, J., Harris, L.D., Krause, K.L., Ward, V.K.(2026) ACS Infect Dis 12: 162-175
- PubMed: 41412823
- DOI: https://doi.org/10.1021/acsinfecdis.5c00680
- Primary Citation of Related Structures:
9PJF, 9PJG, 9PKR - PubMed Abstract:
The cysteine 3C-like proteases (3CL pro ) of caliciviruses, coronaviruses, and picornaviruses are essential for viral replication. In this study, we report the development of potent broad-spectrum peptidomimetic antiviral agents that target the 3CL pro of caliciviruses (NS6), coronaviruses (M pro ), and a picornavirus (3C). Based upon previously reported inhibitors, a small library of compounds was designed, synthesized and tested to identify a core structure, which was then derivatized with a focus upon P3 and P4 positions to afford new inhibitors with improved potency against the respective viral enzymes and enhanced binding as determined by X-ray crystallography. These compounds were tested against a range of viruses in culture, revealing minimal toxicity while exhibiting broad-spectrum potent nanomolar activities against noroviruses and several coronavirus species, including alpha and omicron variants of SARS-CoV-2 and Middle East Respiratory Syndrome virus (MERS).
- School of Chemical Sciences, School of Biological Sciences, The University of Auckland, 23 Symonds Street, 3b Symonds Street, Auckland 1142, New Zealand.
Organizational Affiliation: