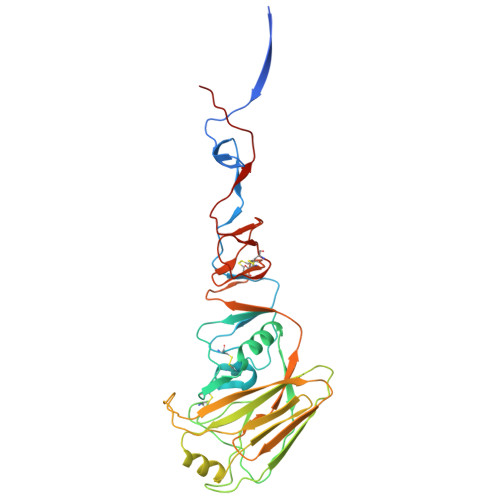
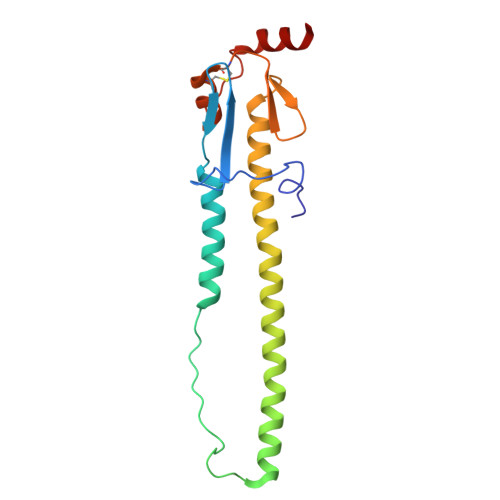
Mechanistic insights into the small-molecule inhibition of influenza A virus entry.
Xu, Y., Anirudhan, V., Gaisina, I.N., Du, H., Alqarni, S., Moore, T.W., Caffrey, M., Manicassamy, B., Zhou, T., Rong, L., Xu, K.(2025) Proc Natl Acad Sci U S A 122: e2503899122-e2503899122
- PubMed: 40802690
- DOI: https://doi.org/10.1073/pnas.2503899122
- Primary Citation of Related Structures:
9ONZ, 9OO1 - PubMed Abstract:
Influenza A virus (IAV) is a zoonotic pathogen responsible for seasonal and pandemic flu. The extensive genetic and antigenic diversity within and between IAV phylogenetic groups presents major challenges for developing universal vaccines and broad-spectrum antiviral therapies. Current interventions provide limited protection due to the virus's high mutation rate and capacity for immune evasion. Recent advancements in viral hemagglutinin (HA)-targeting small-molecule entry inhibitors offer a promising avenue to overcome these limitations. Here, we present structural and functional analyses of two group 2 HA-specific small-molecule inhibitors recently identified by our team. Cryogenic electron microscopy (cryo-EM) structures revealed that these inhibitors bind a conserved pocket within the HA stalk, likely interfering with the conformational rearrangements necessary for membrane fusion and viral entry. Structure-guided mutagenesis confirmed the critical roles of key interacting residues and uncovered distinct resistance profiles between the two compounds, as well as in comparison to Arbidol, a previously reported HA inhibitor. Notably, our structural analysis highlights intrinsic barriers to achieving cross-group inhibition with current small-molecule designs. To address this, we propose an alternative strategy for broadening antiviral coverage. Together, these findings provide mechanistic insights into IAV entry inhibition and a foundation for the rational design of next-generation anti-influenza therapeutics.
- Department of Veterinary Biosciences, The Ohio State University, Columbus, OH 43210.
Organizational Affiliation: