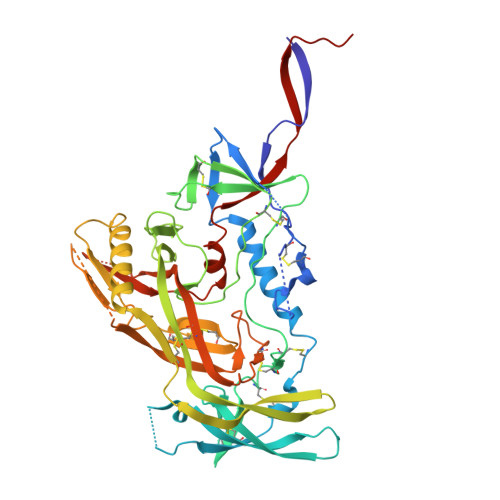
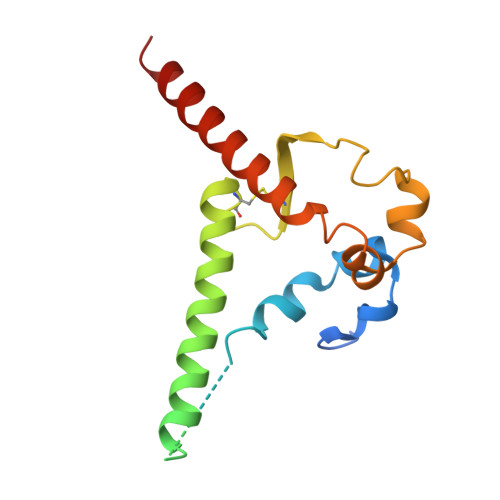
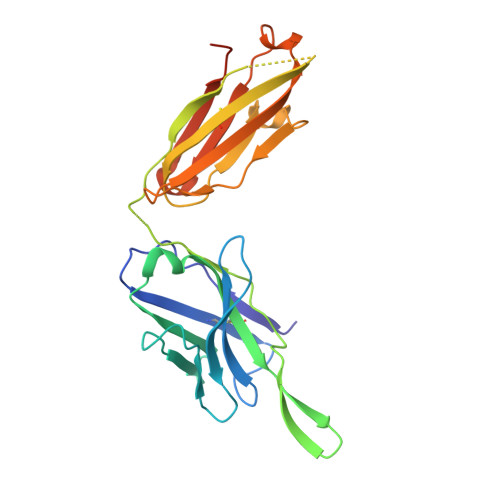
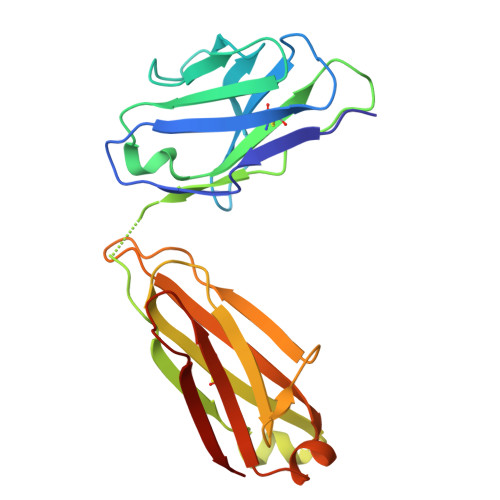
Identification of a broad and potent V3 glycan site bNAb targeting an N332 gp120 glycan-independent epitope.
Gieselmann, L., DeLaitsch, A.T., Rohde, M., Radford, C., Worczinski, J., Momot, A., Ahmadov, E., Burger, J.A., Havenar-Daughton, C., Deshpande, S., Giovannoni, F., Corti, D., Kreer, C., Ercanoglu, M.S., Schommers, P., Georgiev, I.S., West, A.P., Knufer, J., Stumpf, R., Kroidl, A., Geldmacher, C., Maganga, L., William, W., Ntinginya, N.E., Hoelscher, M., Yang, Z., Wei, Q., Renfrow, M., Green, T.J., Novak, J., van Gils, M.J., Gristick, H.B., Gruell, H., Bloom, J.D., Seaman, M.S., Bjorkman, P.J., Klein, F.(2025) bioRxiv
- PubMed: 40964353
- DOI: https://doi.org/10.1101/2025.09.05.674437
- Primary Citation of Related Structures:
9O2Q, 9O2R, 9O2S, 9O2T, 9O2U - PubMed Abstract:
Broadly neutralizing antibodies (bNAbs) against HIV-1 can suppress viremia in vivo and inform vaccine development. Here, we characterized 007, a V3 glycan site bNAb exhibiting high levels of antiviral activity against multiclade pseudovirus panels 1-3 (GeoMean IC 50 = 0.012 μg/mL, breadth = 69%, 217 virus strains) by targeting a N332 gp120 glycan-independent V3 epitope, a site of Env vulnerability to which only weakly neutralizing antibodies had previously been identified. Functional analyses demonstrated distinct binding and neutralization profiles compared to classical V3 glycan site bNAbs. A 007 Fab-Env cryo-EM structure revealed contacts with the V3 324 GD/NIR 327 motif and interactions with N156 gp120 and N301 gp120 glycans. In contrast to classical V3 bNAbs, 007 binding to Env does not depend on the N332 gp120 glycan, rendering it resistant to common escape mutations. Structures of 007 IgG-Env trimer complexes showed two Env trimers crosslinked by three bivalent IgGs, and bivalent 007 IgG was up to ~300-fold more potent than monovalent 007 IgG heterodimer, suggesting a role for avidity in potent neutralization. Finally, in HIV-1 ADA -infected humanized mice, 007 caused transient decline of viremia and overcame classical V3 escape mutations, highlighting 007's potential for HIV-1 prevention, therapy, functional cure, and vaccine design.
- Laboratory of Experimental Immunology, Institute of Virology, Faculty of Medicine and University Hospital Cologne, University of Cologne, 50931 Cologne, Germany.
Organizational Affiliation: