Structure of the ciliary tip central pair reveals the unique role of the microtubule-seam binding protein SPEF1.
Legal, T., Joachimiak, E., Parra, M., Peng, W., Tam, A., Black, C., Guha, M., Nguyen, C.A., Ghanaeian, A., Valente-Paterno, M., Brouhard, G., Gaertig, J., Wloga, D., Bui, K.H.(2025) Curr Biol 35: 3404
- PubMed: 40651469
- DOI: https://doi.org/10.1016/j.cub.2025.06.020
- Primary Citation of Related Structures:
9NTM, 9NW3, 9OT2 - PubMed Abstract:
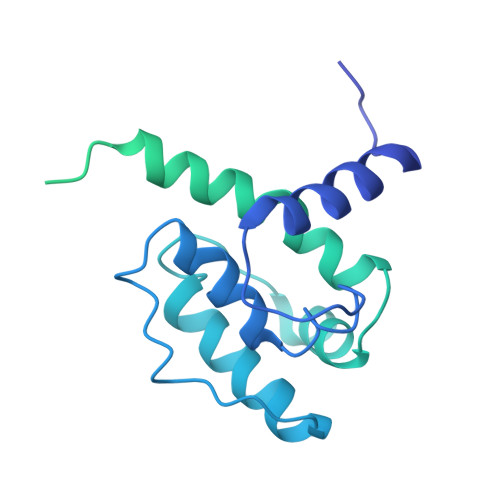
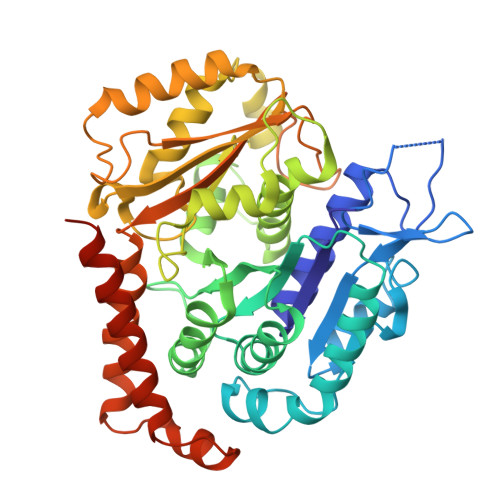
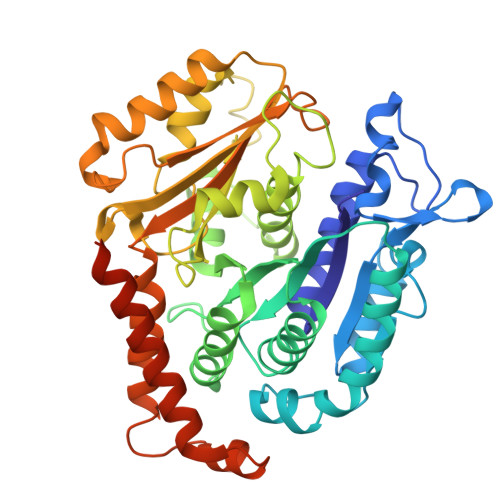
Motile cilia are unique organelles with the ability to move autonomously. The force generated by beating cilia propels cells and moves fluids. The ciliary skeleton is made of peripheral doublet microtubules and a central pair (CP) with a distinct structure at the tip. In this study, we present a high-resolution structure of the CP in the ciliary tip of the ciliate Tetrahymena thermophila and identify several tip proteins that bind and form unique patterns on both microtubules of the tip CP. Two of those proteins that contain tubulin polymerization-promoting protein (TPPP)-like domains, TLP1 and TLP2, bind to high curvature regions of the microtubule. TLP2, which contains two TPPP-like domains, is an unusually long protein that wraps laterally around half a microtubule and forms the bridge between the two microtubules. Moreover, we found that the conserved protein SPEF1 binds to both microtubule seams and crosslinked the two microtubules. In vitro, human SPEF1 binds to the microtubule seam as visualized by cryoelectron tomography and subtomogram averaging. Single-molecule microtubule dynamics assays indicate that SPEF1 stabilizes microtubules in vitro. Together, these data show that the proteins in the tip CP maintain stable microtubule structures and play important roles in maintaining the integrity of the axoneme.
- Department of Anatomy and Cell Biology, McGill University, 3640 Rue University, Montreal, QC H3A 0C7, Canada.
Organizational Affiliation: