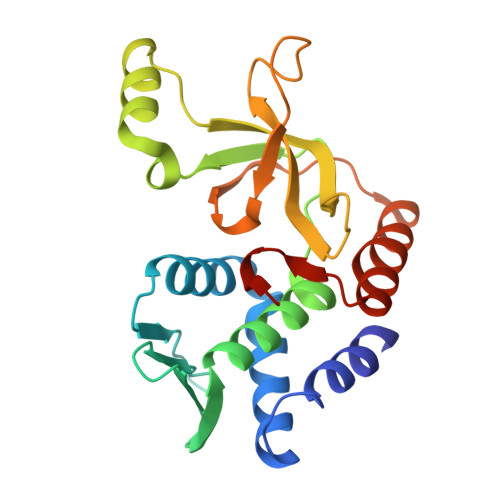
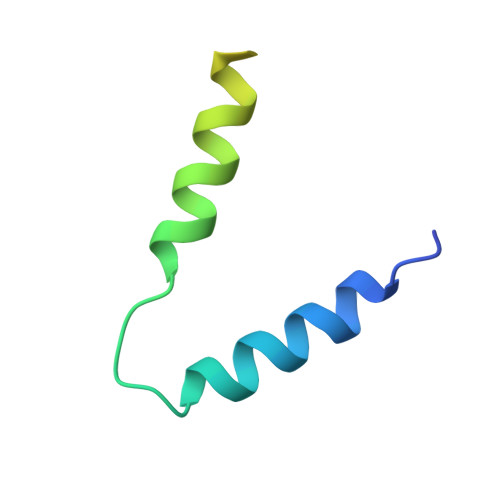
A widespread family of molecular chaperones promotes the intracellular stability of type VIIb secretion system-exported toxins.
Gkragkopoulou, P., Garrett, S.R., Shah, P.Y., Grebenc, D.W., Klein, T.A., Kim, Y., Whitney, J.C.(2025) Proc Natl Acad Sci U S A 122: e2503581122-e2503581122
- PubMed: 40953262
- DOI: https://doi.org/10.1073/pnas.2503581122
- Primary Citation of Related Structures:
9DCR, 9DCS, 9DCT, 9MNO - PubMed Abstract:
To survive in highly competitive environments, bacteria use specialized secretion systems to deliver antibacterial toxins into neighboring cells, thereby inhibiting their growth. In many Gram-positive bacteria, the export of such toxins requires a membrane-bound molecular apparatus known as the type VIIb secretion system (T7SSb). Recently, it was shown that toxin recruitment to the T7SSb requires a physical interaction between a toxin and two or more so-called targeting factors, which harbor key residues required for T7SS-dependent protein export. However, in addition to these targeting factors, some toxins additionally require a protein belonging to the DUF4176 protein family. Here, by examining two toxin-DUF4176 protein pairs, we demonstrate that DUF4176 constitutes a family of toxin-specific molecular chaperones. In addition to being required for toxin stability in producing cells, we find that DUF4176 proteins facilitate toxin export by specifically interacting with a previously uncharacterized intrinsically disordered region found in many T7SS toxins. Using X-ray crystallography, we determine structures of several DUF4176 chaperones in their unbound state, and of a DUF4176 chaperone in complex with the binding site of its cognate toxin. These structures reveal that this binding site consists of a disordered amphipathic α-helix that requires interaction with its cognate chaperone for proper folding. Overall, we have identified a family of secretion system associated molecular chaperones found throughout T7SSb-containing Gram-positive bacteria.
- Michael DeGroote Institute for Infectious Disease Research, McMaster University, Hamilton, ON L8S 4K1, Canada.
Organizational Affiliation: