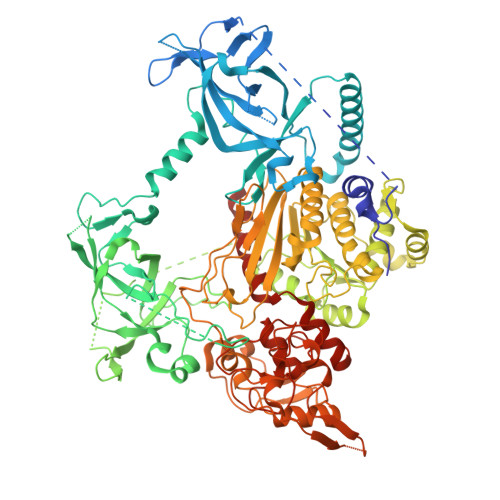
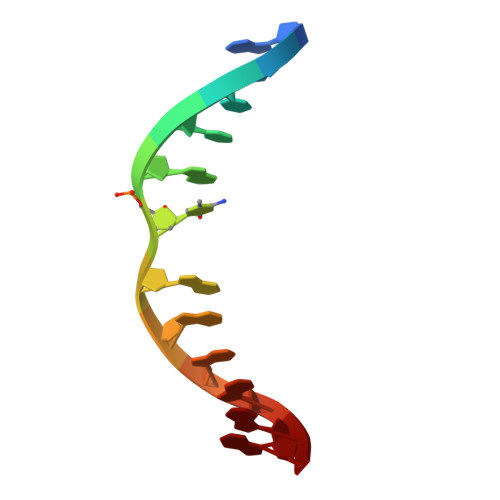
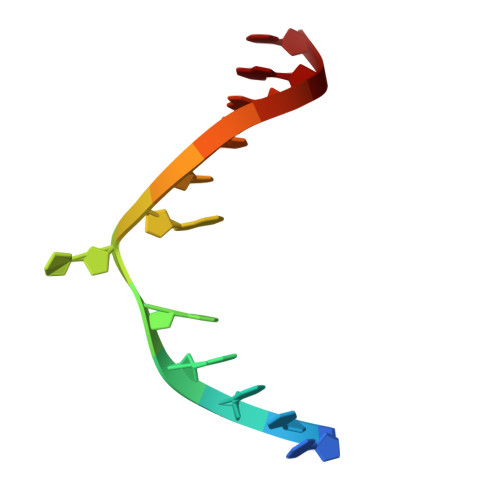
Cryo-EM reveals evolutionarily conserved and distinct structural features of plant CG maintenance methyltransferase MET1.
Kikuchi, A., Nishiyama, A., Chiba, Y., Nakanishi, M., To, T.K., Arita, K.(2025) Nat Commun 16: 8524-8524
- PubMed: 41006297
- DOI: https://doi.org/10.1038/s41467-025-63765-9
- Primary Citation of Related Structures:
9M5U, 9M5X - PubMed Abstract:
DNA methylation is essential for genomic function and transposable element silencing. In plants, DNA methylation occurs in CG, CHG, and CHH contexts (where H = A, T, or C), with the maintenance of CG methylation mediated by the DNA methyltransferase MET1. The molecular mechanism by which MET1 maintains CG methylation, however, remains unclear. Here, we report cryogenic electron microscopy structures of Arabidopsis thaliana MET1. We find that the methyltransferase domain of MET1 specifically methylates hemimethylated DNA in vitro. The structure of MET1 bound to hemimethylated DNA reveals the activation mechanism of MET1 resembling that of mammalian DNMT1. Curiously, the structure of apo-MET1 shows an autoinhibitory state distinct from that of DNMT1, where the RFTS2 domain and the connecting linker inhibit DNA binding. The autoinhibition of MET1 is relieved upon binding of a potential activator, ubiquitinated histone H3. Taken together, our structural analysis demonstrates both conserved and distinct molecular mechanisms regulating CG maintenance methylation in plant and animal DNA methyltransferases.
- Structural Biology Laboratory, Graduate School of Medical Life Science, Yokohama City University, Yokohama, Kanagawa, Japan.
Organizational Affiliation: