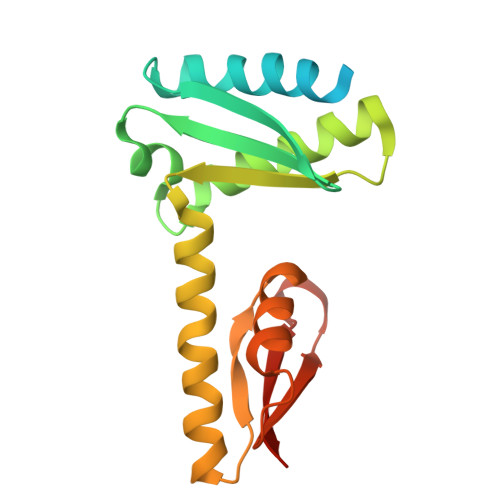
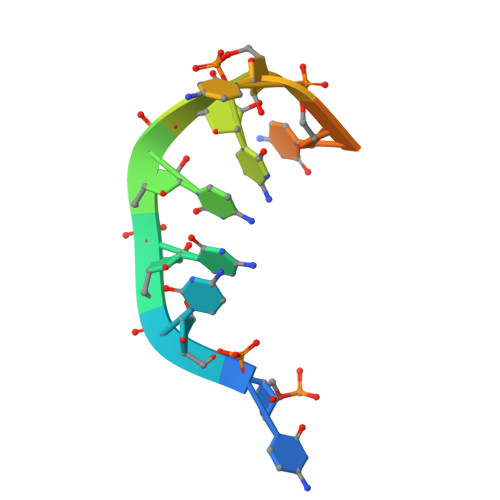
KH-R3H domain cooperation in RNA recognition by the global RNA-binding protein KhpB.
Fukui, K., Murakawa, T., Baba, S., Kumasaka, T., Yano, T.(2025) Nat Commun 16: 8028-8028
- PubMed: 40903470
- DOI: https://doi.org/10.1038/s41467-025-62302-y
- Primary Citation of Related Structures:
9LRG, 9LRI - PubMed Abstract:
KhpB, also known as EloR, is a recently discovered global RNA-binding protein in various pathogenic bacteria that regulates critical cellular processes. KhpB is unique in containing both an R3H domain and a KH domain, which are universal RNA/DNA-binding domains found across various proteins involved in diverse cellular functions. However, the precise roles of these domains in KhpB's RNA-binding mechanism remain unclear, particularly as no structural data of the R3H domain bound to RNA/DNA have been reported for any protein. In this study, we present the crystal structures of both the RNA-free and RNA-bound forms of Thermus thermophilus KhpB dimer. These structures reveal that the KH and R3H domains cooperate to form a composite RNA-binding site capable of binding a single RNA molecule. Notably, the coordinated interaction requires RNA molecules that are at least 7 nucleotides long. This interaction induces conformational changes, including the closure of the RNA-binding cleft between the two domains. The structural data further reveal that KhpB primarily interacts with the phosphate backbone of RNA, while most of the base moieties remain solvent-exposed. These findings provide structural insights into the molecular function of KhpB and shed light on the RNA-binding strategies of other R3H domain-containing proteins.
- Department of Biochemistry, Faculty of Medicine, Osaka Medical and Pharmaceutical University, 2-7 Daigaku-machi, Takatsuki, Osaka, Japan. kenji.fukui@ompu.ac.jp.
Organizational Affiliation: