Structural insights into ligand recognition and G protein preferences across histamine receptors.
Matsuzaki, Y., Sano, F.K., Oshima, H.S., Akasaka, H., Kobayashi, K., Tanaka, T., Itoh, Y., Shihoya, W., Kise, Y., Kusakizako, T., Inoue, A., Nureki, O.(2025) Commun Biol 8: 957-957
- PubMed: 40579541
- DOI: https://doi.org/10.1038/s42003-025-08363-7
- Primary Citation of Related Structures:
9LRB, 9LRC, 9LRD, 9LRE - PubMed Abstract:
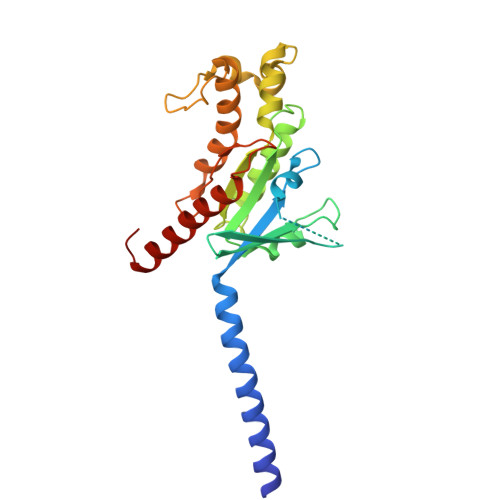
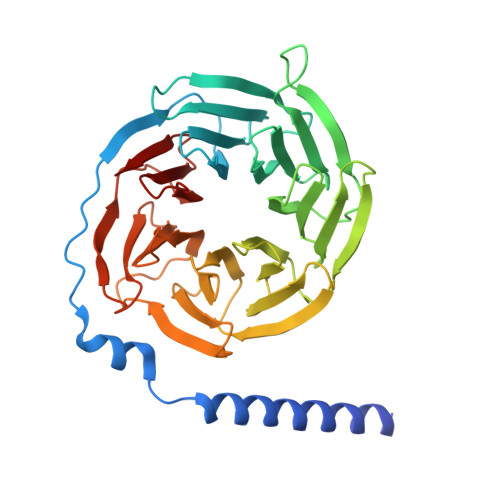
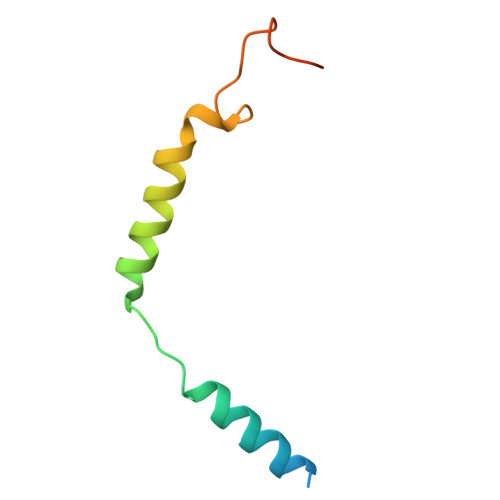
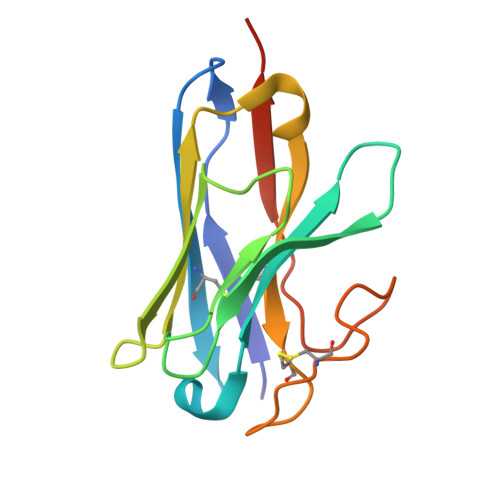
Histamine exerts critical physiological roles by activating four receptor subtypes, each exhibiting a specific G protein preference. Among these, the histamine H 4 receptor (H 4 R) modulates chemotaxis and interferon production through G i protein activation, suggesting its therapeutic potential. Despite its physiological significance, the mechanisms underlying H 4 R signalling and G protein preference across histamine receptors remain poorly understood. Here, we present the cryo-electron microscopy structure of the H 4 R-G i complex, revealing unique mechanisms of histamine recognition and receptor activation. We further solved the structures of the histamine H 1 receptor (H 1 R) bound to the non-canonical G proteins G i and G s . Through a combination of functional and computational analyses, we identified the intracellular loop 2 as a critical determinant of G protein preference in H 1 R and H 4 R. Collectively, our comprehensive study revealed the structural basis for distinct mechanisms of ligand recognition and receptor activation, offering a profound insight into G protein preference across receptor subtypes.
- Department of Biological Sciences, Graduate School of Science, The University of Tokyo, Bunkyo-Ku, Tokyo, Japan.
Organizational Affiliation: