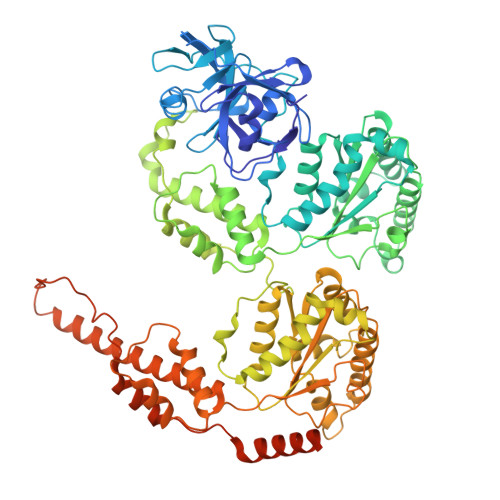
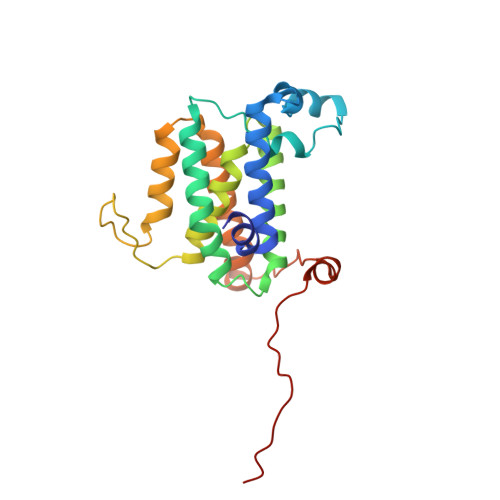
Cryo-EM structure of the human Derlin-1/p97 complex reveals a hexameric channel in ERAD.
Wang, Q., Yao, D., Rao, B., Xia, Y., Li, W., Li, S., Cao, M., Shen, Y., Qin, A., Cao, Y.(2025) Commun Biol 8: 1481-1481
- PubMed: 41107410
- DOI: https://doi.org/10.1038/s42003-025-08880-5
- Primary Citation of Related Structures:
9LLK - PubMed Abstract:
The ER-associated degradation (ERAD) pathway retrotranslocates misfolded proteins from the ER lumen to the cytoplasm for proteasomal degradation. Derlin-1 and p97 are central to this process, forming a canonical 4:6 complex with tetrameric Derlin-1. Using cryo-electron microscopy, we identify a novel human Derlin-1/p97 complex with a 6:6 stoichiometry, where hexameric Derlin-1 assembles as three dimers. This hexameric channel forms a significantly larger trans-ER membrane tunnel, potentially accommodating bulkier substrates. Structural comparisons revealed conformational flexibility in Derlin-1, suggesting the "U"-shaped tetramer may act as an intermediate in hexamer formation. The formation of this hexameric channel is mediated by interactions with p97 and appears dependent on p97's ATPase activity, which provides the driving force for the transition between the tetrameric channel conformation to the intermediate "U"-shaped conformation. These findings highlight the dynamic nature of the Derlin-1/p97 complex and its implications for understanding ERAD retrotranslocation.
- Department of Orthopaedics, Shanghai Key Laboratory of Orthopaedic Implant, Shanghai Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China.
Organizational Affiliation: