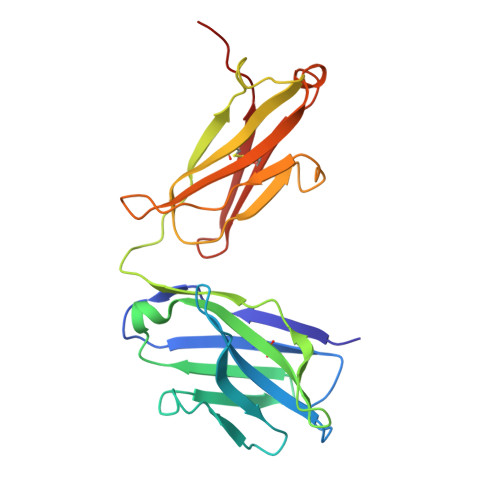
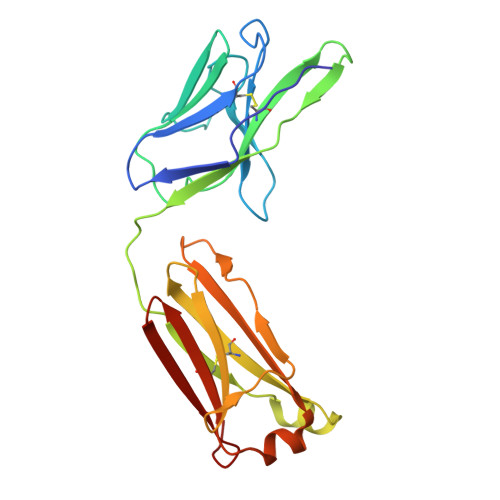
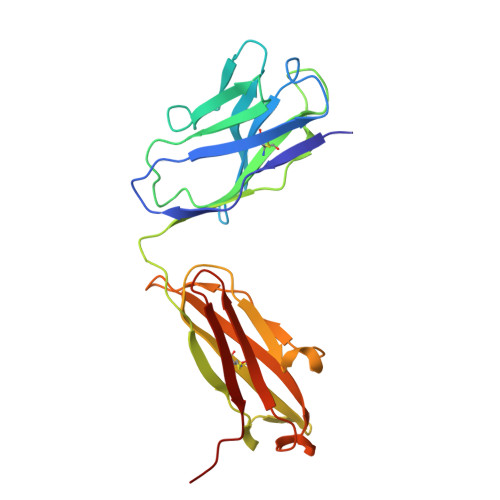
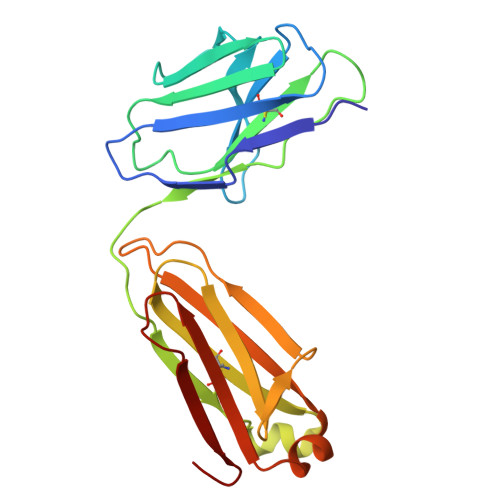
Discovery and structural investigation of Varicella-Zoster virus gE-neutralizing antibodies isolated from a convalescent patient.
Wang, L., Jia, Z., Ye, X., Chen, C., Sun, B., Zhao, X., Zhang, R., Li, Y., Wang, W., Sun, Z., Zhou, L., Ni, Z., Zhang, N., Guo, Y.(2025) Protein Cell 16: 1054-1059
- PubMed: 40583453
- DOI: https://doi.org/10.1093/procel/pwaf051
- Primary Citation of Related Structures:
9JZQ - State Key Laboratory of Medicinal Chemical Biology and College of Life Sciences, Nankai University, Tianjin 300071, China.
Organizational Affiliation: