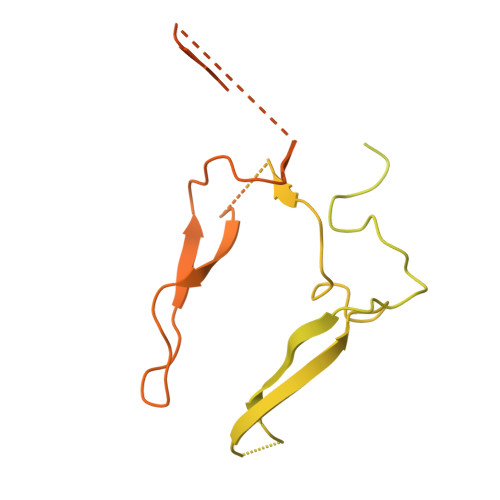
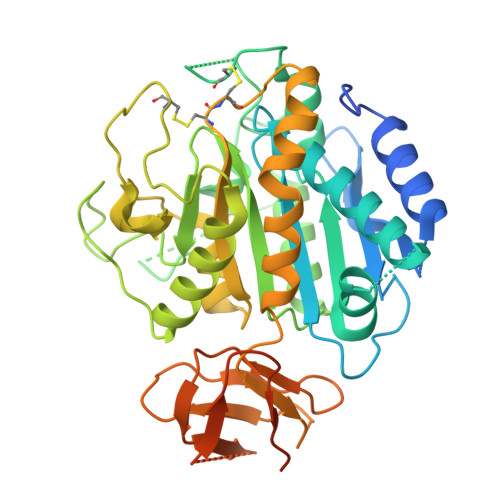
An alternate receptor for adeno-associated viruses.
Dhungel, B.P., Xu, H., Nagarajah, R., Vitale, J., Wong, A.C.H., Gokal, D., Feng, Y., Tabar, M.S., Metierre, C., Parsania, C., Song, X., Wang, G., Su, X.D., Bailey, C.G., Rasko, J.E.J.(2025) Cell 188: 4924
- PubMed: 40664211
- DOI: https://doi.org/10.1016/j.cell.2025.06.026
- Primary Citation of Related Structures:
9J6Z, 9J7K, 9J7L - PubMed Abstract:
Systemic gene therapy using adeno-associated virus (AAV) vectors is approved for the treatment of several genetic disorders, but challenges and toxicities associated with high vector doses remain. We report an alternate receptor for AAV (AAVR2, carboxypeptidase D [CPD]), which is distinct from the multi-serotype AAV receptor (AAVR). AAVR2 enables the transduction of clade E AAVs, including AAV8, and determines an exclusive AAVR-independent transduction pathway for AAV11 and AAV12. We characterized direct binding between the AAV8 capsid and AAVR2 by cryo-electron microscopy (cryo-EM) and identified contact residues. We observed that AAV8 directly binds to the carboxypeptidase-like domain 1 of AAVR2 via its variable region VIII and demonstrated that AAV capsids that lack AAVR2 binding can be bioengineered to engage with AAVR2. Finally, we overexpressed a minimal functional AAVR2 to enhance AAV transduction in vivo. Our study provides insights into AAV biology and clinically deployable solutions to reduce dose-related toxicities associated with AAV vectors.
- School of Medical Sciences, Faculty of Medicine & Health, The University of Sydney, Camperdown, NSW 2006, Australia; Cancer & Gene Regulation Laboratory Centenary Institute, The University of Sydney, Camperdown, NSW 2050, Australia; Centre for Rare Diseases & Gene Therapy Centenary Institute, The University of Sydney, Camperdown, NSW 2050, Australia.
Organizational Affiliation: