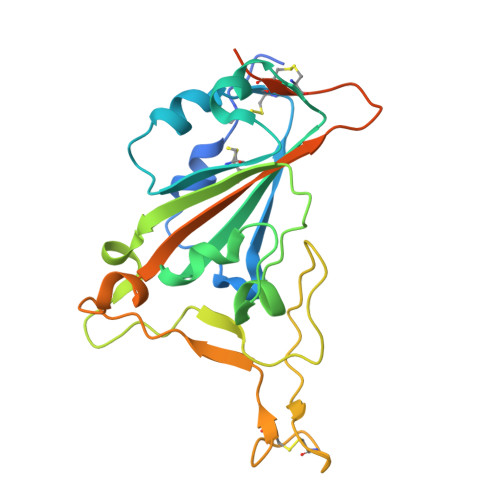
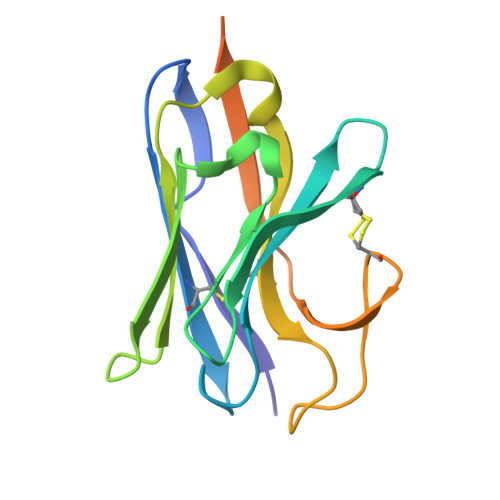
Ultra-potent RBM-specific single-domain antibody broadly neutralizes multiple SARS-CoV-2 variants with picomolar activity.
Shcheblyakov, D.V., Favorskaya, I.A., Dolzhikova, I.V., Korobkova, A.I., Alekseeva, I.A., Esmagambetov, I.B., Voronina, O.L., Tukhvatulin, A.I., Zubkova, O.V., Derkaev, A.A., Ryabova, E.I., Iliukhina, A.A., Zorkov, I.D., Grousova, D.M., Reshetnikov, D.A., Ryzhova, N.N., Ermolova, E.I., Kunda, M.S., Matyuta, I.O., Boyko, K.M., Popov, V.O., Logunov, D.Y., Sluchanko, N.N., Gintsburg, A.L.(2025) Int J Biol Macromol 319: 145386-145386
- PubMed: 40543774
- DOI: https://doi.org/10.1016/j.ijbiomac.2025.145386
- Primary Citation of Related Structures:
9IQP - PubMed Abstract:
SARS-CoV-2 infection remains a cause of severe illness in high-risk individuals, with few antiviral agents currently available. The emergence of new SARS-CoV-2 variants accumulating an increasing number of mutations significantly challenges the development of effective therapeutics. We describe broadly neutralizing single-domain antibody 1p1B10 having picomolar activity against both previously circulating SARS-CoV-2 variants, including Wuhan D614G, Alpha, Beta, Gamma, Delta and Omicron BA.1, BA.2, BA.5, and more recent variants - XBB.1, XBB.1.5, XBB.1.9, XBB.1.16, JN.1 and KS.1. We explained this broad activity by solving a high-resolution crystal structure of the S-protein RBD complex with the 1p1B10 antibody. The RBD/1p1B10 interface is unaffected by accumulated mutations and substantially overlaps with the RBD/ACE2 interface. 1p1B10 acts through binding to RBD both in open and closed conformations and blocking SARS-CoV-2 attachment to cells via direct competition with the ACE2 receptor. Therapeutic 1p1B10-Fc administration at a low dose of 1 mg/kg substantially reduced viral load in the lungs of Syrian hamsters after challenge with evolutionary distant SARS-CoV-2 variants and completely protected hACE2 mice against lethal SARS-CoV-2 infection. Overall, the findings make 1p1B10 a promising candidate for etiotropic treatment of COVID-19.
- Federal State Budget Institution "National Research Center for Epidemiology and Microbiology Named after Honorary Academician N.F. Gamaleya" of the Ministry of Health of the Russian Federation, 123098 Moscow, Russia. Electronic address: sdmitryv@yahoo.com.
Organizational Affiliation: