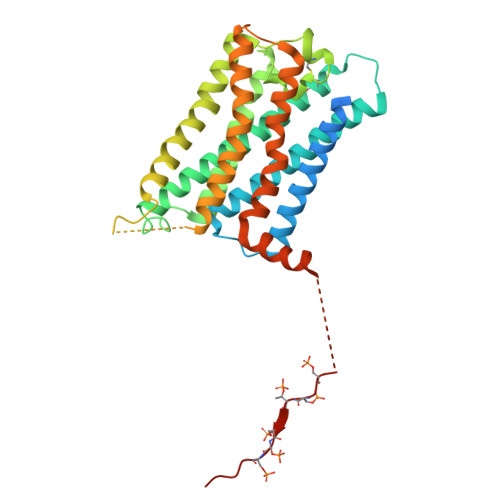
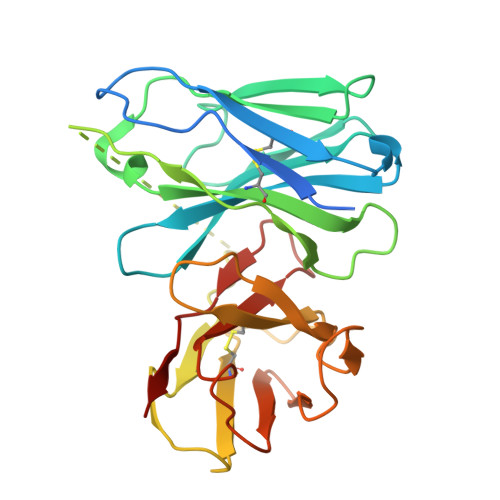
Constitutive arrestin recruitment by orphan GPR52 via an atypical binding mode.
Lin, X., Wei, X., Pu, N., Wang, L., Zhang, Z., Li, C., Yue, Y., Liu, J., Tan, Q., Sun, Q., Xu, F.(2025) Cell Res 35: 913-916
- PubMed: 40813798
- DOI: https://doi.org/10.1038/s41422-025-01165-w
- Primary Citation of Related Structures:
9IJR, 9IJS - iHuman Institute, ShanghaiTech University, Shanghai, China. linxi@shanghaitech.edu.cn.
Organizational Affiliation: