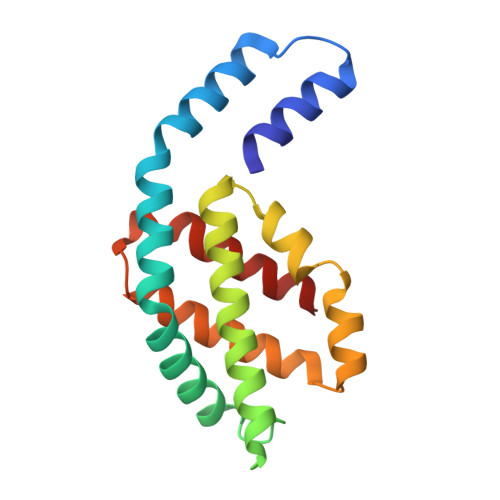
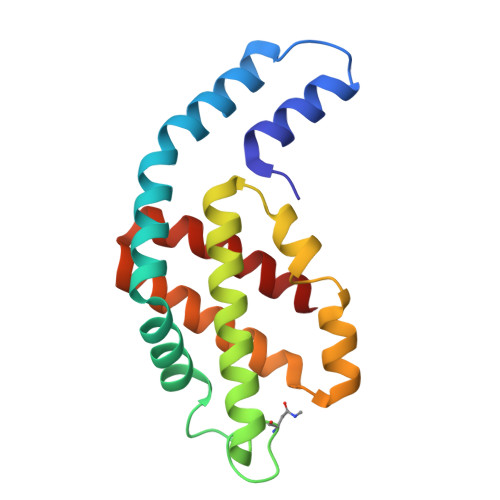
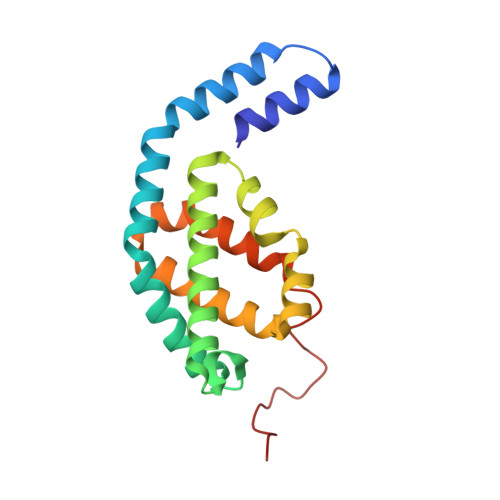
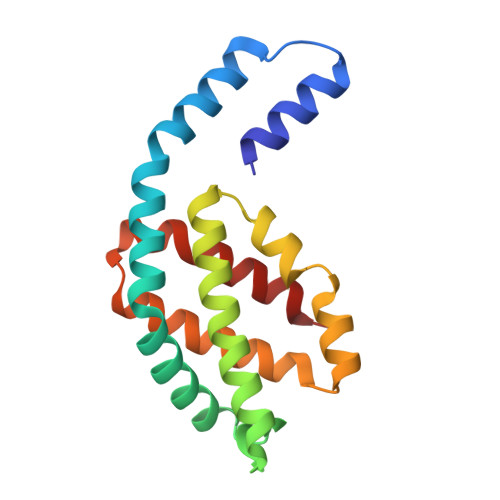
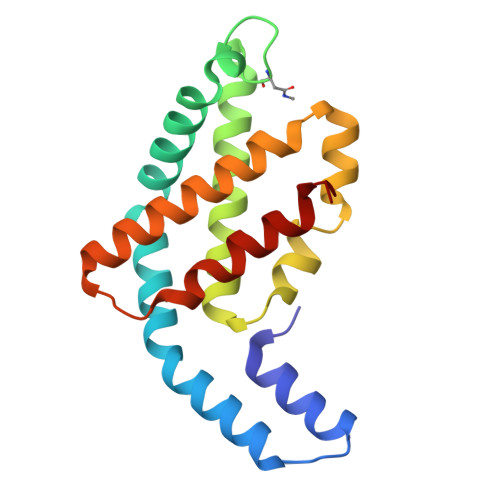
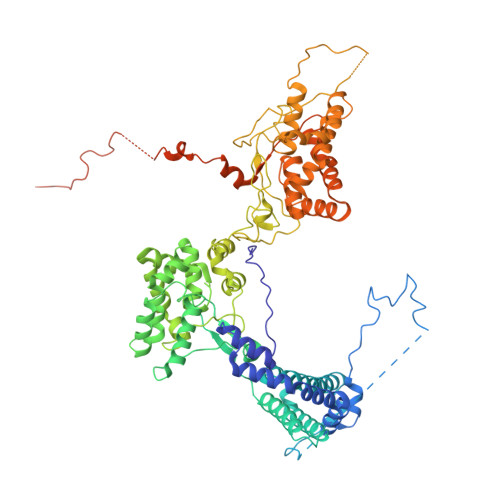
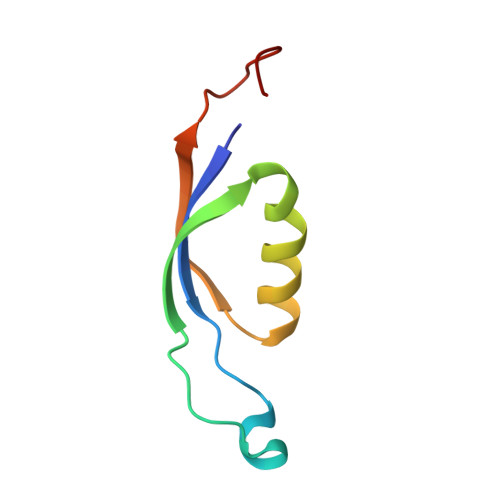
Structure of a stripped-down and tuned-up far-red phycobilisome.
Consoli, G., Leong, H.F., Davis, G.A., Richardson, T., McInnes, A., Murray, J.W., Fantuzzi, A., Rutherford, A.W.(2025) Commun Biol 8: 907-907
- PubMed: 40494956
- DOI: https://doi.org/10.1038/s42003-025-08326-y
- Primary Citation of Related Structures:
9I1R - PubMed Abstract:
A diverse subset of cyanobacteria can transiently modify their photosynthetic machinery during far-red light photoacclimation to drive photosynthesis with less energetic photons (700 nm-800 nm). To achieve this, all the main light-driven components of the photosynthetic apparatus, including their allophycocyanin antenna, are replaced with red-shifted paralogues. Recent studies based on the structure of an incomplete complex provided some insights into the tuning of the far-red phycobiliproteins. Here, we solved the structure of the intact bicylindrical allophycocyanin complex from the cyanobacterium Chroococcidiopsis thermalis PCC 7203 at a resolution of 2.51 Å determined by Cryo-electron microscopy single particle analysis. A comparison between conserved structural features in far-red and white light allophycocyanin cores provides insight on the evolutionary adaptations needed to optimize excitation energy transfer in the far-red light adapted photosynthetic apparatus. The reduction in antenna size in far-red photosynthesis suggests a need to optimize membrane packing to increase the number of photosystems and tune the ratio between chlorophyll f molecules and bilin pigments, while the wider spread in the absorption range of the bilins suggests faster and more efficient excitation energy transfer to far-red Photosystem II by limiting backflow of excitation from the reaction centres to the far-red bilin pigments.
- Department of Life Sciences, Imperial College, London, UK.
Organizational Affiliation: