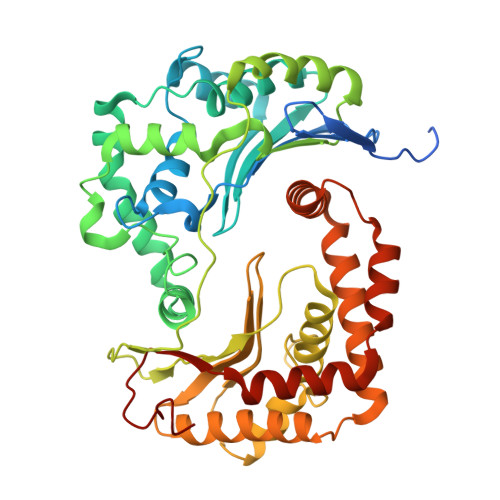
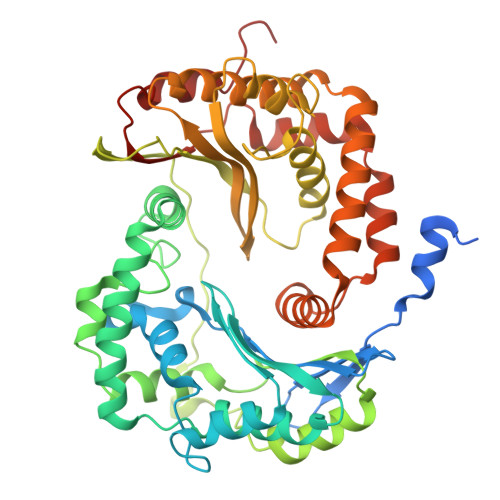
Structural basis for late maturation steps of mitochondrial respiratory chain complex IV within the human respirasome.
Nguyen, M.D., Sierra-Magro, A., Singh, V., Khawaja, A., Timon-Gomez, A., Barrientos, A., Rorbach, J.(2026) Nat Commun
- PubMed: 41519940
- DOI: https://doi.org/10.1038/s41467-025-68274-3
- Primary Citation of Related Structures:
9HZL, 9I6F, 9I7U, 9TI4 - PubMed Abstract:
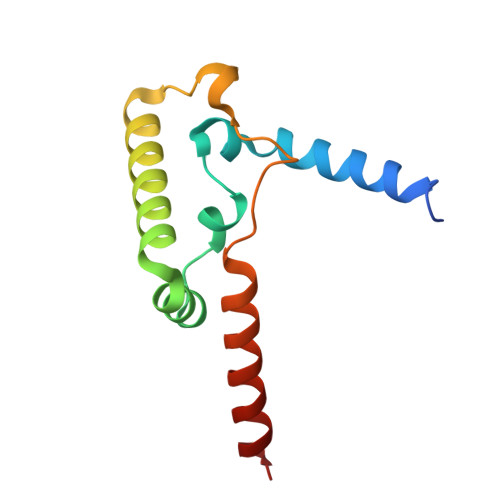
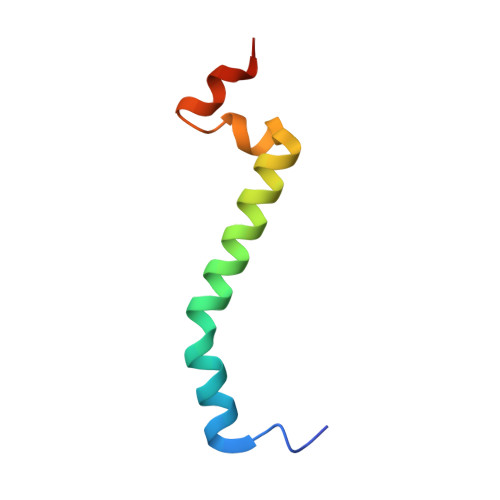
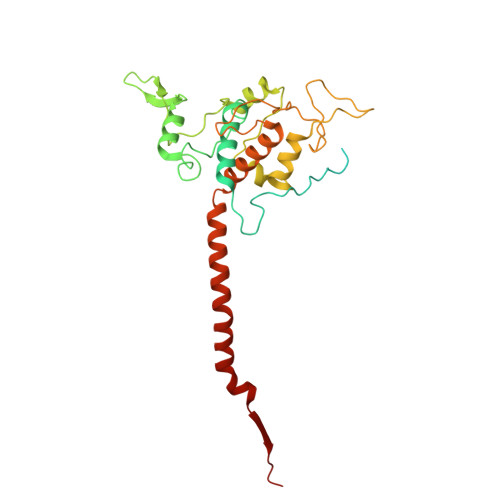
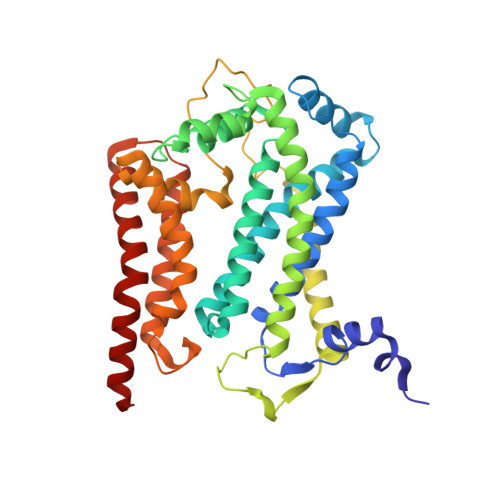
The mitochondrial respiratory chain comprises four multimeric complexes (CI-CIV) that drive oxidative phosphorylation by transferring electrons to oxygen and generating the proton gradient required for ATP synthesis. These complexes can associate into supercomplexes (SCs), such as the CI + CIII₂ + CIV respirasome, but how SCs form, by joining preassembled complexes or by engaging partially assembled intermediates, remains unresolved. Here, we use cryo-electron microscopy to determine high-resolution structures of native human CI + CIII₂ + CIV late-assembly intermediates. Together with biochemical analyses, these structures show that respirasome biogenesis concludes with the final maturation of CIV while it is associated with fully assembled CI and CIII₂. We identify HIGD2A as a placeholder factor within isolated and supercomplexed CIV that is replaced by subunit NDUFA4 during the last step of CIV and respirasome assembly. This mechanism suggests that placeholders such as HIGD2A act as molecular timers, preventing premature incorporation of NDUFA4 or its isoforms and ensuring the orderly progression of pre-SC particles into functional respirasomes. Since defects in CIV assembly, including NDUFA4 deficiencies, cause severe encephalomyopathies and neurodegenerative disorders, understanding the molecular architecture and assembly pathways of isolated and supercomplexed CIV offers insight into the pathogenic mechanisms underlying these conditions.
- Department of Medical Biochemistry and Biophysics, Karolinska Institutet, Stockholm, Sweden.
Organizational Affiliation: