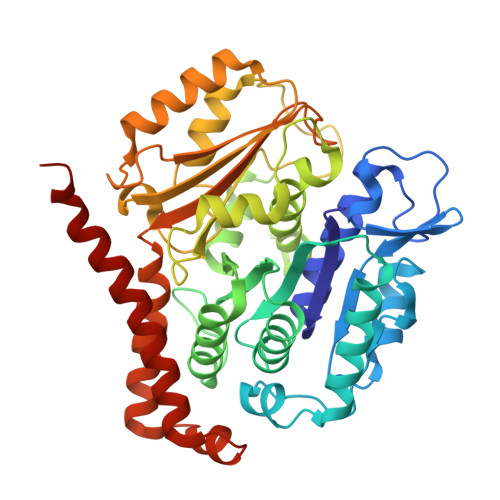
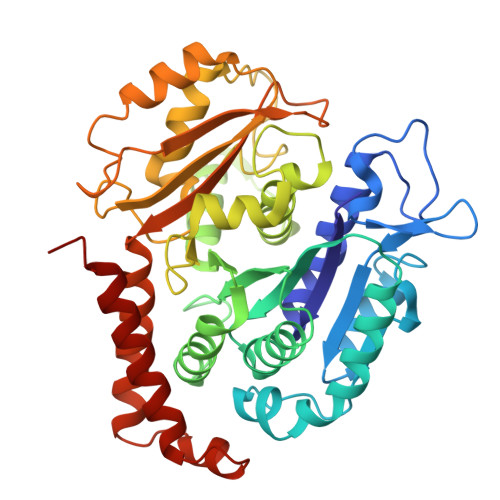
Microtubules in Asgard archaea.
Wollweber, F., Xu, J., Ponce-Toledo, R.I., Marxer, F., Rodrigues-Oliveira, T., Possnecker, A., Luo, Z.H., Malit, J.J.L., Kokhanovska, A., Wieczorek, M., Schleper, C., Pilhofer, M.(2025) Cell 188: 2451
- PubMed: 40120574
- DOI: https://doi.org/10.1016/j.cell.2025.02.027
- Primary Citation of Related Structures:
9F6T, 9F6U, 9F6V, 9HXC - PubMed Abstract:
Microtubules are a hallmark of eukaryotes. Archaeal and bacterial homologs of tubulins typically form homopolymers and non-tubular superstructures. The origin of heterodimeric tubulins assembling into microtubules remains unclear. Here, we report the discovery of microtubule-forming tubulins in Asgard archaea, the closest known relatives of eukaryotes. These Asgard tubulins (AtubA/B) are closely related to eukaryotic α/β-tubulins and the enigmatic bacterial tubulins BtubA/B. Proteomics of Candidatus Lokiarchaeum ossiferum showed that AtubA/B were highly expressed. Cryoelectron microscopy structures demonstrate that AtubA/B form eukaryote-like heterodimers, which assembled into 5-protofilament bona fide microtubules in vitro. The additional paralog AtubB2 lacks a nucleotide-binding site and competitively displaced AtubB. These AtubA/B2 heterodimers polymerized into 7-protofilament non-canonical microtubules. In a sub-population of Ca. Lokiarchaeum ossiferum cells, cryo-tomography revealed tubular structures, while expansion microscopy identified AtubA/B cytoskeletal assemblies. Our findings suggest a pre-eukaryotic origin of microtubules and provide a framework for understanding the fundamental principles of microtubule assembly.
- Department of Biology, Institute of Molecular Biology & Biophysics, Eidgenössische Technische Hochschule Zürich, Otto-Stern-Weg 5, 8093 Zürich, Switzerland.
Organizational Affiliation: