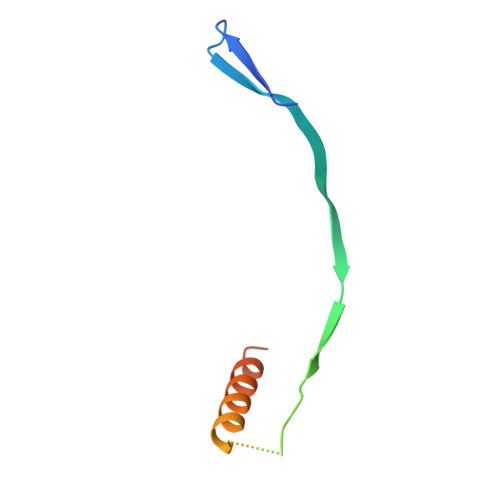
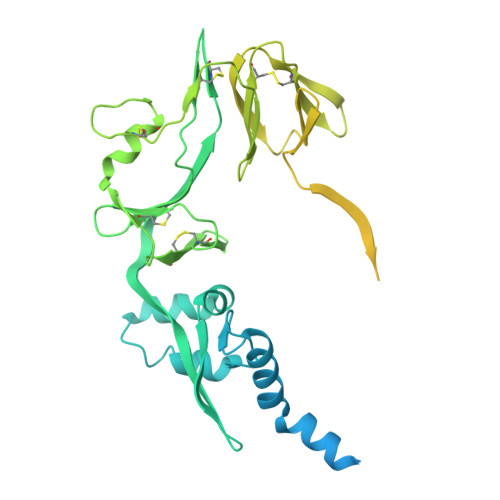
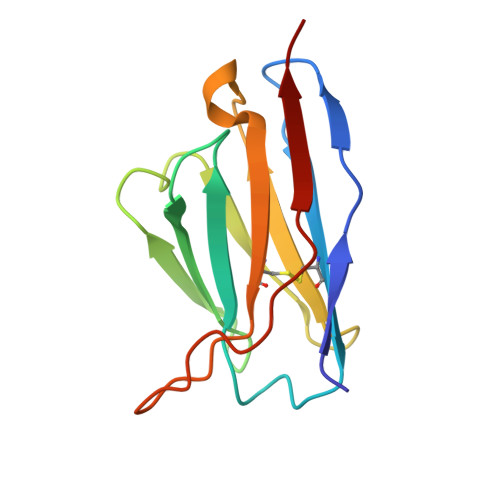
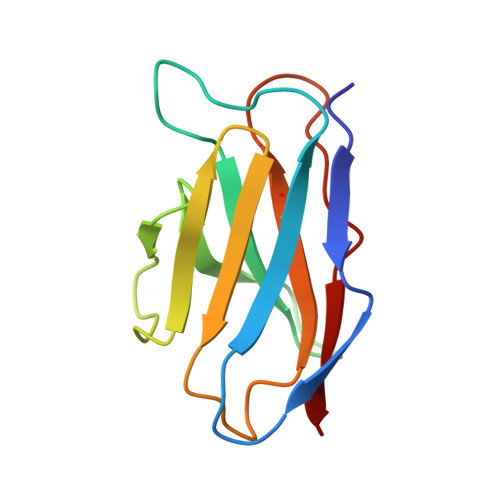
Structural Basis for Postfusion-Specific Binding to the Respiratory Syncytial Virus F Protein by the Canonical Antigenic Site I Antibody 131-2a.
Peng, W., Siborova, M., Wu, X., Du, W., Schulte, D., Pronker, M.F., de Haan, C.A.M., Snijder, J.(2025) ACS Infect Dis 11: 2357-2366
- PubMed: 40693554
- DOI: https://doi.org/10.1021/acsinfecdis.5c00368
- Primary Citation of Related Structures:
9HVW - PubMed Abstract:
The respiratory syncytial virus (RSV) fusion (F) protein is a major target of antiviral antibodies following natural infection or vaccination and is responsible for mediating fusion between the viral envelope and the host membrane. The fusion process is driven by a large-scale conformational change in F, switching irreversibly from the metastable prefusion state to the stable postfusion conformation. Previous research has identified six distinct antigenic sites in RSV-F, termed sites Ø, I, II, III, IV, and V. Of these, only antigenic site I is fully specific to the postfusion conformation of F. A monoclonal antibody 131-2a that specifically targets postfusion F has been widely used as a research tool to probe for postfusion F and to define antigenic site I in serological studies, yet its sequence and precise epitope have remained unknown. Here, we use mass spectrometry-based de novo sequencing of 131-2a to reverse engineer a recombinant product and study the epitope to define antigenic site I with molecular detail, revealing the structural basis for the antibody's specificity toward postfusion RSV-F.
- Biomolecular Mass Spectrometry and Proteomics, Bijvoet Center for Biomolecular Research and Utrecht Institute of Pharmaceutical Sciences, Utrecht University, Padualaan 8, Utrecht 3584CH, the Netherlands.
Organizational Affiliation: