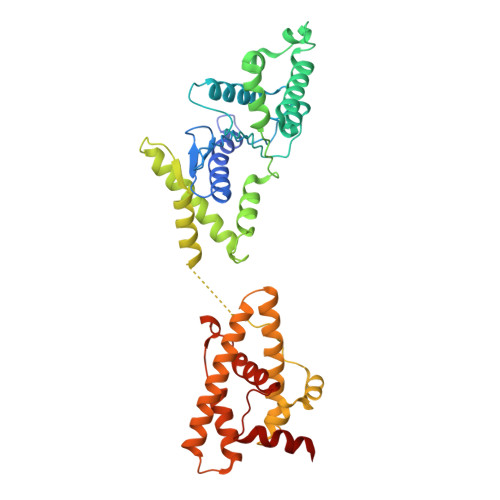
Structure and activation mechanism of a Lamassu phage and plasmid defense system.
Li, Y., Adams, D.W., Liu, H.W., Shaw, S.J., Uchikawa, E., Jaskolska, M., Stutzmann, S., Righi, L., Szczelkun, M.D., Blokesch, M., Gruber, S.(2025) Nat Struct Mol Biol 32: 2503-2516
- PubMed: 41087596
- DOI: https://doi.org/10.1038/s41594-025-01677-4
- Primary Citation of Related Structures:
9HQU, 9HQX, 9HR5 - PubMed Abstract:
Lamassu is a diverse family of defense systems that protect bacteria, including seventh-pandemic strains of Vibrio cholerae, against both plasmids and phage infection. During phage infection, Lamassu targets essential cellular processes, thereby halting phage propagation by terminating the infected host. The mechanisms by which Lamassu effectors are activated when needed and otherwise suppressed are unknown. Here we present structures of a Lamassu defense system from Salmonella enterica. We show that an oligomerization domain of the nuclease effector subunit, LmuA, is sequestered by two tightly folded SMC-like LmuB protomers and LmuC. Upon activation, liberated LmuA assembles into homotetramers, in which two of four nuclease domains are brought into proximity to create an active site capable of cleaving DNA. We propose that tetramer formation is likely a one-way switch that establishes a threshold to limit potential spontaneous activation and cell death. Our findings reveal a mechanism of cellular defense, involving liberation and oligomerization of immune effectors, and shed light on how Lamassu systems balance potent immune responses with self-preservation.
- Department of Fundamental Microbiology (DMF), Faculty of Biology and Medicine (FBM), University of Lausanne (UNIL), Lausanne, Switzerland.
Organizational Affiliation: