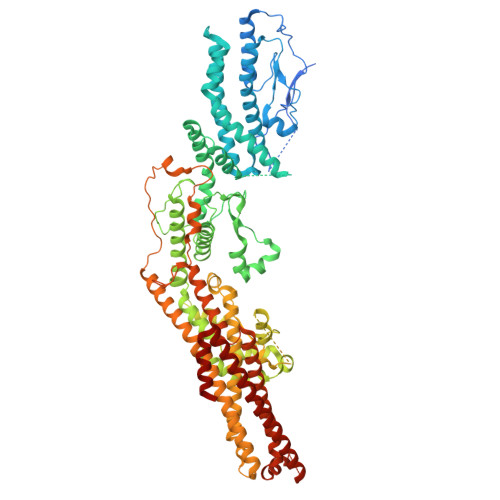
Structural insights into the interplay between microtubule polymerases, gamma-tubulin complexes and their receptors.
Zheng, A., Vermeulen, B.J.A., Wurtz, M., Neuner, A., Lubbehusen, N., Mayer, M.P., Schiebel, E., Pfeffer, S.(2025) Nat Commun 16: 402-402
- PubMed: 39757296
- DOI: https://doi.org/10.1038/s41467-024-55778-7
- Primary Citation of Related Structures:
9H9P, 9H9Q, 9H9R - PubMed Abstract:
The γ-tubulin ring complex (γ-TuRC) is a structural template for controlled nucleation of microtubules from α/β-tubulin heterodimers. At the cytoplasmic side of the yeast spindle pole body, the CM1-containing receptor protein Spc72 promotes γ-TuRC assembly from seven γ-tubulin small complexes (γ-TuSCs) and recruits the microtubule polymerase Stu2, yet their molecular interplay remains unclear. Here, we determine the cryo-EM structure of the Candida albicans cytoplasmic nucleation unit at 3.6 Å resolution, revealing how the γ-TuRC is assembled and conformationally primed for microtubule nucleation by the dimerised Spc72 CM1 motif. Two coiled-coil regions of Spc72 interact with the conserved C-terminal α-helix of Stu2 and thereby position the α/β-tubulin-binding TOG domains of Stu2 in the vicinity of the microtubule assembly site. Collectively, we reveal the function of CM1 motifs in γ-TuSC oligomerisation and the recruitment of microtubule polymerases to the γ-TuRC.
- Zentrum für Molekulare Biologie der Universität Heidelberg (ZMBH), Heidelberg, Germany.
Organizational Affiliation: