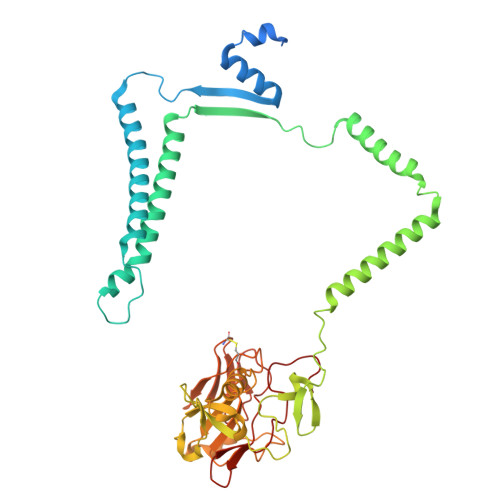
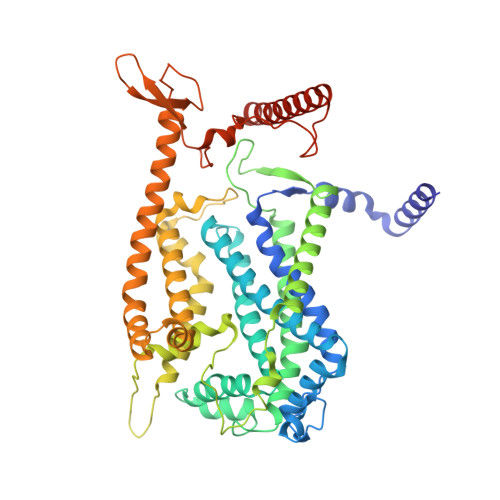
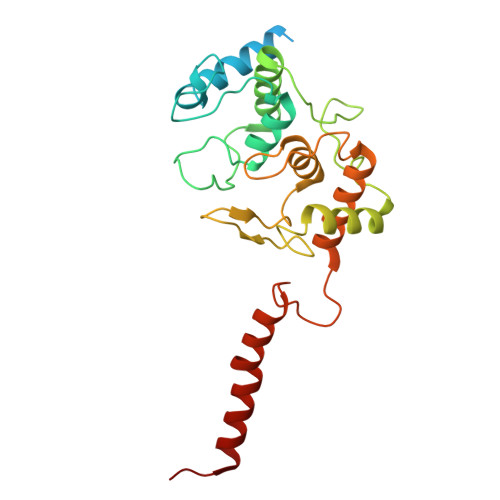
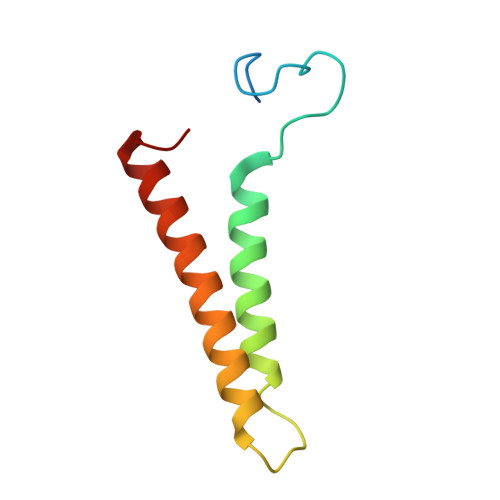
Structural and mechanistic study of a novel inhibitor analogue of M. tuberculosis cytochrome bc 1 :aa 3 .
Verma, A.K., Kim, R.Q., Lamprecht, D.A., Aguilar-Perez, C., Wong, S., Veziris, N., Aubry, A., Bartolome-Nebreda, J.M., Carbajo, R.J., Wetzel, J., Lamers, M.H.(2025) NPJ Drug Discov 2: 6-6
- PubMed: 40191462
- DOI: https://doi.org/10.1038/s44386-025-00008-3
- Primary Citation of Related Structures:
9GY6 - PubMed Abstract:
Drug-resistant tuberculosis (TB) continues to challenge treatment options, necessitating the exploration of new compounds of novel targets. The mycobacterial respiratory complex cytochrome bc 1 :aa 3 has emerged as a promising target, exemplified by the success of first-in-class inhibitor Q203 in phase 2 clinical trials. However, to fully exploit the potential of this target and to identify the best-in-class inhibitor more compounds need evaluation. Here, we introduce JNJ-2901, a novel Q203 analogue, that demonstrates activity against multidrug-resistant M. tuberculosis clinical strains at sub-nanomolar concentration and 4-log reduction in bacterial burden in a mouse model of TB infection. Inhibitory studies on purified enzymes validate the nanomolar inhibitions observed in mycobacterial cells. Additionally, cryo-EM structure analysis of cytochrome bc 1 :aa 3 bound to JNJ-2901 reveals the binding pocket at the menaquinol oxidation site (Qp), akin to other substate analogue inhibitors like Q203 and TB47. Validation of the binding site is further achieved by generating and isolating the JNJ-2901 resistant mutations in M. tuberculosis , followed by purification and resistance analysis of the resistant cytochrome bc 1 :aa 3 complex. Our comprehensive work lays the foundation for further clinical validations of JNJ-2901.
- Department of Cell and Chemical Biology, Leiden University Medical Center, Einthovenweg 20, 2333 ZC Leiden, The Netherlands.
Organizational Affiliation: