Recognition and remodelling of nucleosomes and hexasomes by the human INO80 complex
Aggarwal, P., Sharma, M., Hopfner, K.P.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| RuvB-like 1 | A [auth C], D [auth A], E [auth B] | 456 | Homo sapiens | Mutation(s): 0 Gene Names: RUVBL1, INO80H, NMP238, TIP49, TIP49A EC: 3.6.4.12 | 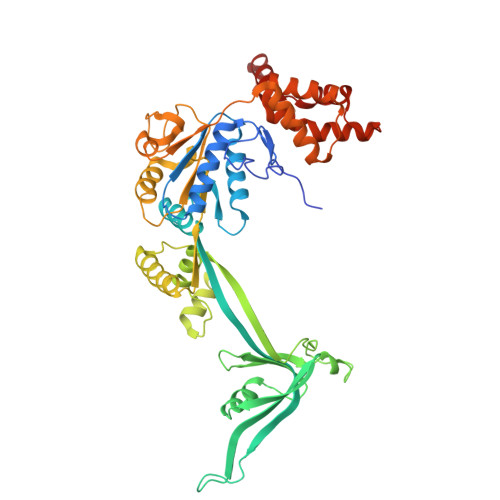 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9Y265 (Homo sapiens) Explore Q9Y265 Go to UniProtKB: Q9Y265 | |||||
PHAROS: Q9Y265 GTEx: ENSG00000175792 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9Y265 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| RuvB-like 2 | B [auth E], F [auth D], G [auth F] | 463 | Homo sapiens | Mutation(s): 0 Gene Names: RUVBL2, INO80J, TIP48, TIP49B, CGI-46 EC: 3.6.4.12 | 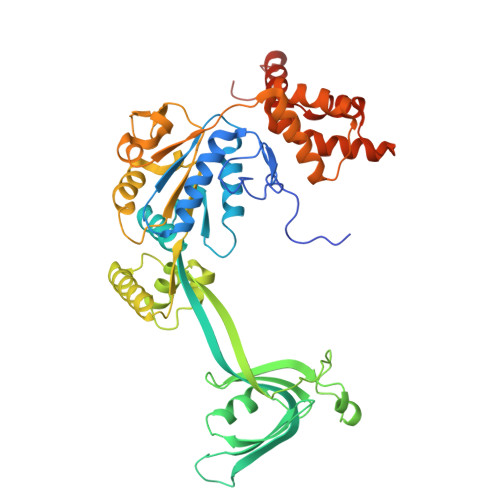 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9Y230 (Homo sapiens) Explore Q9Y230 Go to UniProtKB: Q9Y230 | |||||
PHAROS: Q9Y230 GTEx: ENSG00000183207 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9Y230 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| INO80 complex subunit C | C [auth I] | 192 | Homo sapiens | Mutation(s): 0 Gene Names: INO80C, C18orf37 | 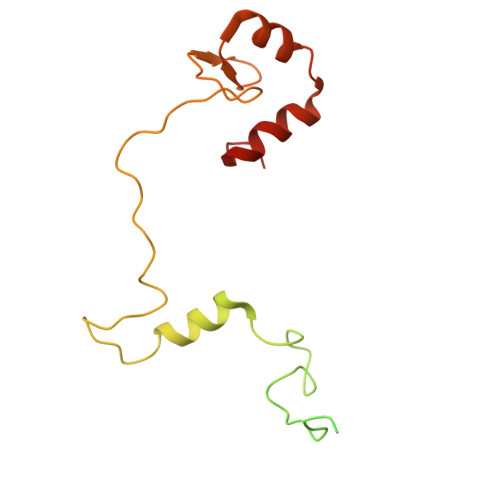 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q6PI98 (Homo sapiens) Explore Q6PI98 Go to UniProtKB: Q6PI98 | |||||
PHAROS: Q6PI98 GTEx: ENSG00000153391 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q6PI98 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Chromatin-remodeling ATPase INO80 | H [auth G] | 1,556 | Homo sapiens | Mutation(s): 0 Gene Names: INO80, INO80A, INOC1, KIAA1259 EC: 3.6.4 | 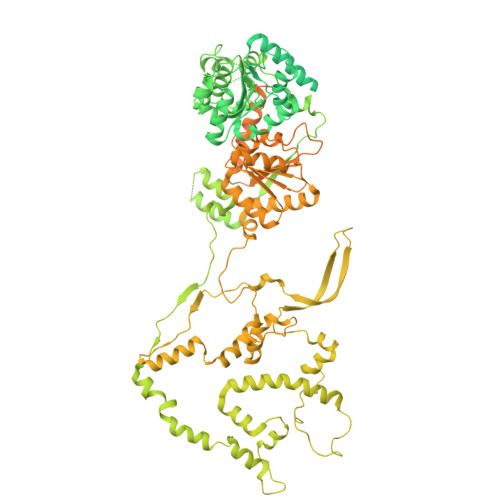 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9ULG1 (Homo sapiens) Explore Q9ULG1 Go to UniProtKB: Q9ULG1 | |||||
PHAROS: Q9ULG1 GTEx: ENSG00000128908 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9ULG1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| INO80 complex subunit B | I [auth H] | 356 | Homo sapiens | Mutation(s): 0 Gene Names: INO80B, HMGA1L4, PAPA1, ZNHIT4 |  |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9C086 (Homo sapiens) Explore Q9C086 Go to UniProtKB: Q9C086 | |||||
GTEx: ENSG00000115274 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9C086 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Actin-related protein 5 | 607 | Homo sapiens | Mutation(s): 0 Gene Names: ACTR5, ARP5 |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9H9F9 (Homo sapiens) Explore Q9H9F9 Go to UniProtKB: Q9H9F9 | |||||
PHAROS: Q9H9F9 GTEx: ENSG00000101442 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9H9F9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 9 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Histone H3.1 | 136 | Homo sapiens | Mutation(s): 0 Gene Names: |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P68431 (Homo sapiens) Explore P68431 Go to UniProtKB: P68431 | |||||
PHAROS: P68431 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P68431 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 10 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Histone H4 | 102 | Homo sapiens | Mutation(s): 0 Gene Names: |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P62805 (Homo sapiens) Explore P62805 Go to UniProtKB: P62805 | |||||
PHAROS: P62805 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62805 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 11 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Histone H2A.Z | 127 | Homo sapiens | Mutation(s): 0 Gene Names: H2AZ1, H2AFZ, H2AZ |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P0C0S5 (Homo sapiens) Explore P0C0S5 Go to UniProtKB: P0C0S5 | |||||
PHAROS: P0C0S5 GTEx: ENSG00000164032 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0C0S5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 12 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Histone H2B type 2-E | 125 | Homo sapiens | Mutation(s): 0 Gene Names: H2BC21, H2BFQ, HIST2H2BE |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q16778 (Homo sapiens) Explore Q16778 Go to UniProtKB: Q16778 | |||||
PHAROS: Q16778 GTEx: ENSG00000184678 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q16778 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 13 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Histone H2A.Z | 127 | Homo sapiens | Mutation(s): 0 Gene Names: H2AZ1, H2AFZ, H2AZ |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P0C0S5 (Homo sapiens) Explore P0C0S5 Go to UniProtKB: P0C0S5 | |||||
PHAROS: P0C0S5 GTEx: ENSG00000164032 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0C0S5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| Nucleosomal DNA Strand 1 (152-MER) | 152 | synthetic construct |  | ||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 8 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| Nucleosomal DNA Strand 2 (152-MER) | 152 | synthetic construct |  | ||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ATP Query on ATP | W [auth A] | ADENOSINE-5'-TRIPHOSPHATE C10 H16 N5 O13 P3 ZKHQWZAMYRWXGA-KQYNXXCUSA-N |  | ||
| ADP Query on ADP | AA [auth G] CA [auth J] U [auth C] V [auth E] X [auth B] | ADENOSINE-5'-DIPHOSPHATE C10 H15 N5 O10 P2 XTWYTFMLZFPYCI-KQYNXXCUSA-N |  | ||
| ZN Query on ZN | BA [auth H] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | REFMAC | 5.8.0425 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| European Research Council (ERC) | European Union | 833613 INO3D |
| German Research Foundation (DFG) | Germany | HO 2489/9-1 |