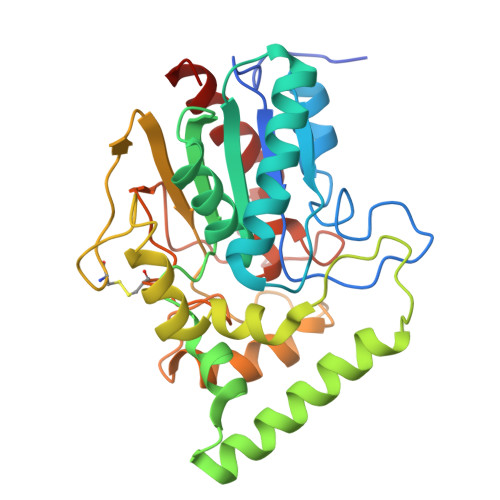
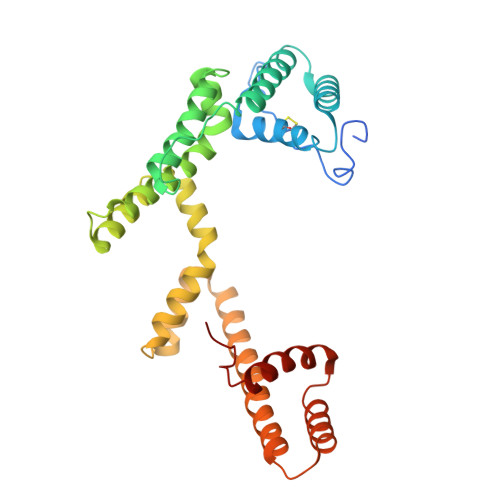
Architecture of an embracing lipase-foldase complex of the type II secretion system of Acinetobacter baumannii.
de Oliveira Silva, Y.R., Contreras-Martel, C., Rodrigues de Melo, R., Zanphorlin, L.M., Trindade, D.M., Dessen, A.(2025) Structure 33: 601-612.e4
- PubMed: 39904335
- DOI: https://doi.org/10.1016/j.str.2024.12.022
- Primary Citation of Related Structures:
9G8U - PubMed Abstract:
Acinetobacter baumannii is a major human pathogen responsible for a growing number of multi-antibiotic-resistant infections, and of critical priority for the World Health Organization (WHO). A. baumannii employs a type II secretion system (T2SS) to secrete toxins extracellularly to enable cytotoxicity and colonization. Lipase LipA, secreted by the A. baumannii T2SS, is required for virulence and fitness, and in the periplasm is maintained in an active state by its essential foldase, LipB. Here we report that LipA is able to recognize lipids of different chain lengths at extremes of pH and temperature, thanks to its stabilization by LipB through an extended, highly helical "embrace." A vast bioinformatic analysis indicates that LipB-like foldases are widespread over numerous proteobacteria, and thus the extended foldase architecture shown here could be widespread. These results provide new insight into A. baumannii's adaptability as a pathogen in different environments and could facilitate the development of novel antibacterials.
- Brazilian Biosciences National Laboratory (LNBio), CNPEM, Campinas São Paulo 13084-971, Brazil; Departamento de Genética, Evolução, Microbiologia e Imunologia, Instituto de Biologia, Universidade Estadual de Campinas (UNICAMP), Campinas, São Paulo 13083-970, Brazil.
Organizational Affiliation: