Optimal Stapling of a Helical Peptide-Foldamer Hybrid Using a C-Terminal 4-Mercaptoproline Enhances Protein Surface Recognition and Cellular Activity.
Neuville, M., Bourgeais, M., Buratto, J., Saragaglia, C., Li, B., Galeano-Otero, I., Mauran, L., Varajao, L., Goudreau, S.R., Kauffmann, B., Thinon, E., Pasco, M., Khatib, A.M., Guichard, G.(2025) Chemistry 31: e202403330-e202403330
- PubMed: 40014761
- DOI: https://doi.org/10.1002/chem.202403330
- Primary Citation of Related Structures:
9FQL, 9GFK - PubMed Abstract:
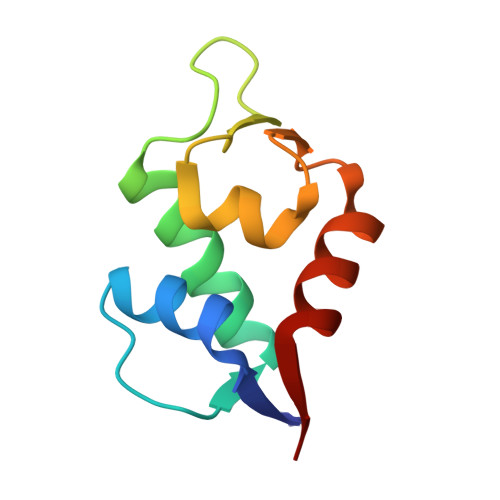
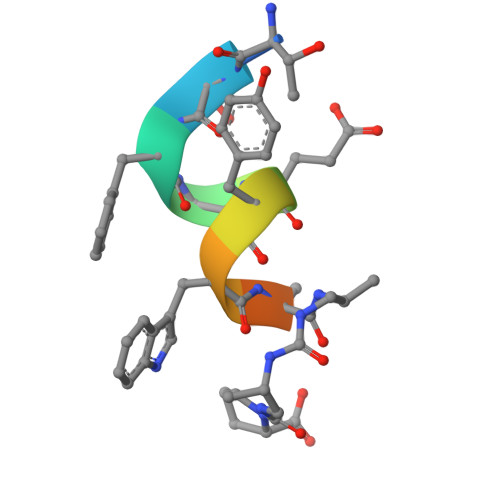
Structural analysis of a co-crystal of a helically-folded peptide-foldamer hybrid in complex with hDM2 E3 ubiquitin ligase, revealed a unique orientation for the C-terminal proline with the pyrrolidine ring pointing backwards in the sequence, and suggested new opportunities for macrocyclization. In particular, we found that the C-terminal prolyl residue could be replaced by its (2S,4S)-4-mercaptoprolyl analogue for optimal bisthioether crosslinking with a cysteine residue installed at position 4 in the sequence. The resulting i,i+7 stapled peptide-foldamer is a high-affinity binder to hDM2, is cell permeable and restores the p53 signalling pathway in p53wt cancer cells. The co-crystal structure of hDM2 and the stapled peptide-foldamer hybrid was determined at 1.84 Å, fully validating the original design and further highlighting the potential of cis-4-mercaptoproline in the context of peptide and foldamer stapling.
- Univ. Bordeaux, CNRS, Bordeaux INP, CBMN, UMR 5248, IECB, 2 rue Robert Escarpit, F-33600, Pessac, France.
Organizational Affiliation: