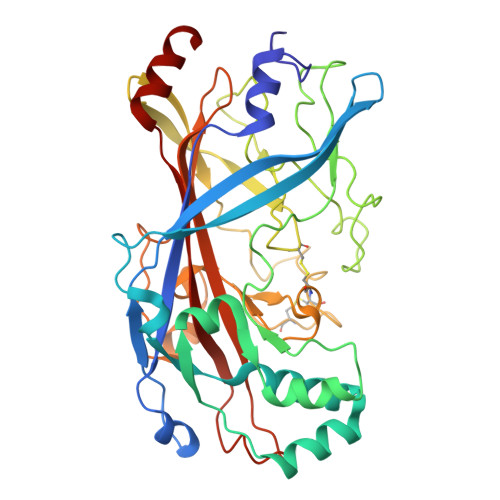
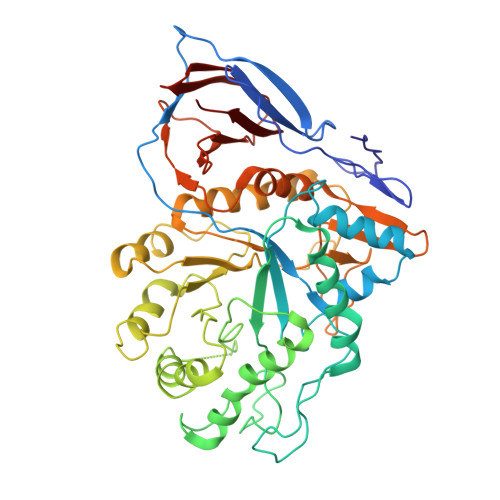
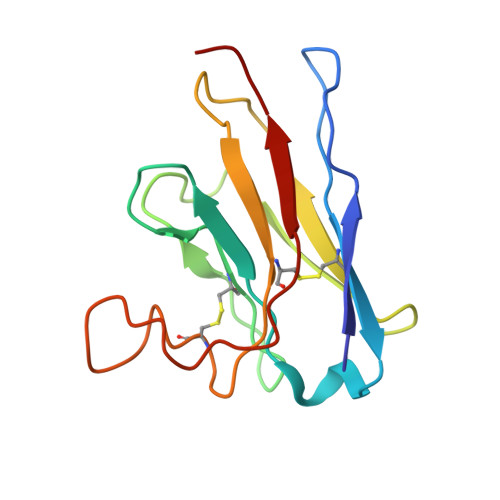
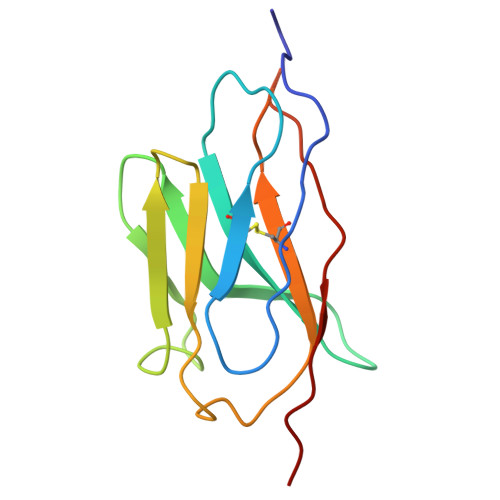
Cryo-TEM structure of beta-glucocerebrosidase in complex with its transporter LIMP-2.
Dobert, J.P., Schafer, J.H., Dal Maso, T., Ravindran, P., Huard, D.J.E., Socher, E., Schildmeyer, L.A., Lieberman, R.L., Versees, W., Moeller, A., Zunke, F., Arnold, P.(2025) Nat Commun 16: 3074-3074
- PubMed: 40159502
- DOI: https://doi.org/10.1038/s41467-025-58340-1
- Primary Citation of Related Structures:
9FJF - PubMed Abstract:
Targeting proteins to their final cellular destination requires transport mechanisms and nearly all lysosomal enzymes reach the lysosome via the mannose-6-phosphate receptor pathway. One of the few known exceptions is the enzyme β-glucocerebrosidase (GCase) that requires the lysosomal integral membrane protein type-2 (LIMP-2) as a proprietary lysosomal transporter. Genetic variations in the GCase encoding gene GBA1 cause Gaucher's disease (GD) and present the highest genetic risk factor to develop Parkinson's disease (PD). Activators targeting GCase emerge as a promising therapeutic approach to treat GD and PD, with pre-clinical and clinical trials ongoing. In this study, we resolve the complex of GCase and LIMP-2 using cryo-electron microscopy with the aid of an engineered LIMP-2 shuttle and two GCase-targeted pro-macrobodies. We identify helix 5 and helix 7 of LIMP-2 to interact with a binding pocket in GCase, forming a mostly hydrophobic interaction interface supported by one essential salt bridge. Understanding the interplay of GCase and LIMP-2 on a structural level is crucial to identify potential activation sites and conceptualizing novel therapeutic approaches targeting GCase. Here, we unveil the protein structure of a mannose-6-phosphate-independent lysosomal transport complex and provide fundamental knowledge for translational clinical research to overcome GD and PD.
- Department of Molecular Neurology, University Hospital Erlangen, Friedrich-Alexander-University Erlangen-Nürnberg (FAU), Erlangen, Germany.
Organizational Affiliation: