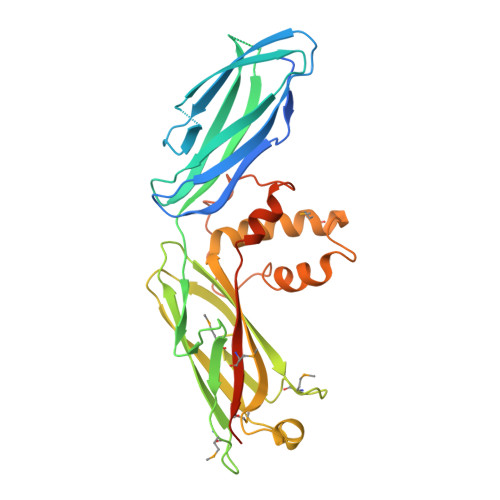
The conserved Spd-2/CEP192 domain adopts a unique protein fold to promote centrosome scaffold assembly.
Hu, L., Wainman, A., Andreeva, A., Apizi, M., Alvarez-Rodrigo, I., Wong, S.S., Saurya, S., Sheppard, D., Cottee, M., Johnson, S., Lea, S.M., Raff, J.W., van Breugel, M., Feng, Z.(2025) Sci Adv 11: eadr5744-eadr5744
- PubMed: 40106572
- DOI: https://doi.org/10.1126/sciadv.adr5744
- Primary Citation of Related Structures:
9C72, 9FH8, 9FU8 - PubMed Abstract:
Centrosomes form when centrioles assemble pericentriolar material (PCM) around themselves. Spd-2/CEP192 proteins, defined by a conserved "Spd-2 domain" (SP2D) comprising two closely spaced AspM-Spd-2-Hydin (ASH) domains, play a critical role in centrosome assembly. Here, we show that the SP2D does not target Drosophila Spd-2 to centrosomes but rather promotes PCM scaffold assembly. Crystal structures of the human and honeybee SP2D reveal an unusual "extended cradle" structure mediated by a conserved interaction interface between the two ASH domains. Mutations predicted to perturb this interface, including a human mutation associated with male infertility and Mosaic Variegated Aneuploidy, disrupt PCM scaffold assembly in flies. The SP2D is monomeric in solution, but the Drosophila SP2D can form higher-order oligomers upon phosphorylation by PLK1 (Polo-like kinase 1). Crystal-packing interactions and AlphaFold predictions suggest how SP2Ds might self-assemble, and mutations associated with one such potential dimerization interface markedly perturb SP2D oligomerization in vitro and PCM scaffold assembly in vivo.
- State Key Laboratory of Genetic Engineering, School of Life Sciences, Fudan University, Shanghai, China.
Organizational Affiliation: