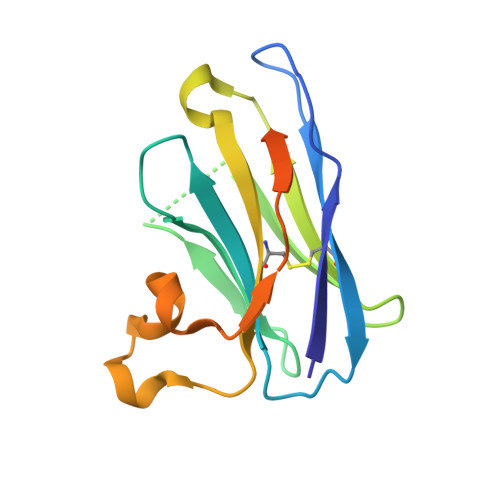
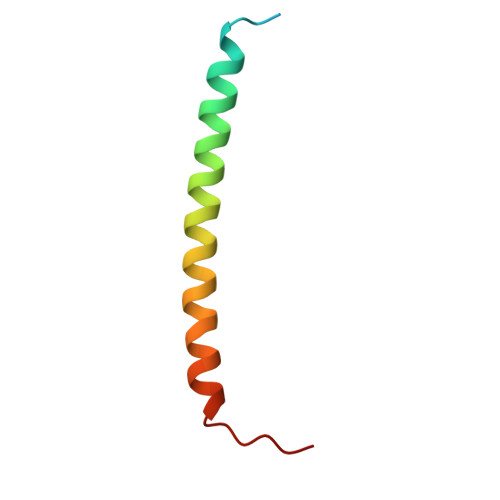
Ultrapotent SARS coronavirus-neutralizing single-domain antibodies that clamp the spike at its base.
De Cae, S., Van Molle, I., van Schie, L., Shoemaker, S.R., Deckers, J., Debeuf, N., Lameire, S., Nerinckx, W., Roose, K., Fijalkowska, D., Devos, S., De Smet, A.S., Marchan, J.C.Z., Venneman, T., Sedeyn, K., Mujanovic, L., Ballegeer, M., Vanheerswynghels, M., De Wolf, C., Demol, H., Zuallaert, J., Vanhaverbeke, P., Ghassabeh, G.H., Lonigro, C., Bockstal, V., Rinaldi, M., Abdelnabi, R., Neyts, J., Marqusee, S., Lambrecht, B.N., Callewaert, N., Remaut, H., Saelens, X., Schepens, B.(2025) Nat Commun 16: 5040-5040
- PubMed: 40447603
- DOI: https://doi.org/10.1038/s41467-025-60250-1
- Primary Citation of Related Structures:
9FCM - PubMed Abstract:
Therapeutic monoclonal antibodies can prevent severe disease in SARS-CoV-2 exposed individuals. However, currently circulating virus variants have evolved to gain significant resistance to nearly all neutralizing human immune system-derived therapeutic monoclonal antibodies that had previously been emergency-authorized for use in the clinic. Here, we describe the discovery of a panel of single-domain antibodies (VHHs) directed against the spike protein S2 subunit that broadly neutralize SARS-CoV-1 and -2 with unusually high potency. One of these VHHs tightly clamps the spike's monomers at a highly conserved, quaternary epitope in the membrane proximal part of the trimeric Heptad Repeat 2 (HR2) coiled-coil, thereby locking the HR2 in its prefusion conformation. Low dose systemic administration of a VHH-human IgG1 Fc fusion prevented SARS-CoV-2 infection in two animal models. Pseudovirus escape selection experiments demonstrate that the very rare escape variants are rendered almost non-infectious. This VHH-based antibody with a highly potent mechanism of antiviral action forms the basis for a new class of pan-sarbecovirus neutralizing biologics, which are currently under development. In addition, the unique quaternary binding mode of the VHHs to the prefusion HR2 could be exploited for other class I fusion proteins.
- VIB Center for Medical Biotechnology, VIB, Ghent, Belgium.
Organizational Affiliation: