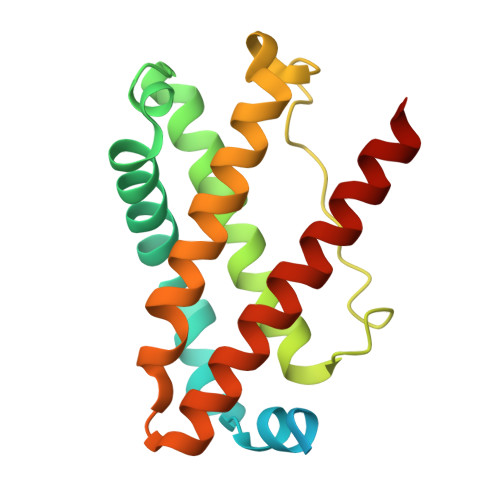
Domain architecture of the Mycobacterium tuberculosis MabR ( Rv2242 ), a member of the PucR transcription factor family.
Megalizzi, V., Tanina, A., Grosse, C., Mirgaux, M., Legrand, P., Dias Mirandela, G., Wohlkonig, A., Bifani, P., Wintjens, R.(2024) Heliyon 10: e40494-e40494
- PubMed: 39641026
- DOI: https://doi.org/10.1016/j.heliyon.2024.e40494
- Primary Citation of Related Structures:
9F80, 9FB1 - PubMed Abstract:
MabR ( Rv2242 ), a PucR-type transcription factor, plays a crucial role in regulating mycolic acid biosynthesis in Mycobacterium tuberculosis . To understand its regulatory mechanisms, we determined the crystal structures of its N-terminal and C-terminal domains. The N-terminal domain adopts a globin-like fold, while the C-terminal domain comprises an α/β GGDEF domain and an all-α effector domain with a helix-turn-helix DNA-binding motif. This unique domain combination is specific to Actinomycetes . Biochemical and computational studies suggest that full-length MabR forms both dimeric and tetrameric assemblies in solution. Structural analysis revealed two distinct dimerization interfaces within the N- and C-terminal domains, further supporting a tetrameric organization. These findings provide valuable insights into the domain architecture, oligomeric state, and potential regulatory mechanisms of MabR.
- Unit of Microbiology, Bioorganic and Macromolecular Chemistry, Department of Research in Drug Development, Faculty of Pharmacy, Université Libre de Bruxelles, Belgium.
Organizational Affiliation: