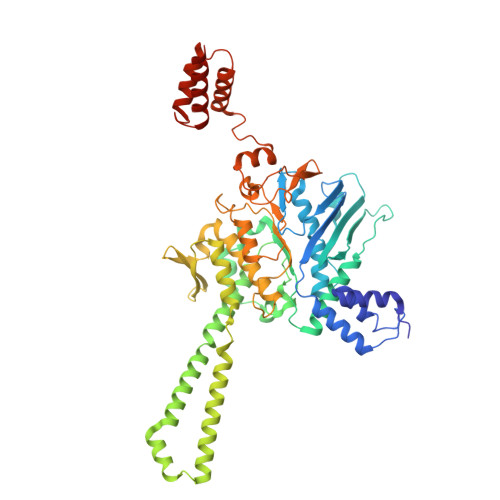
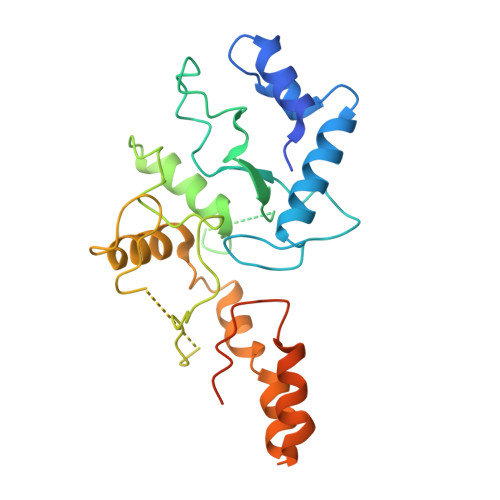
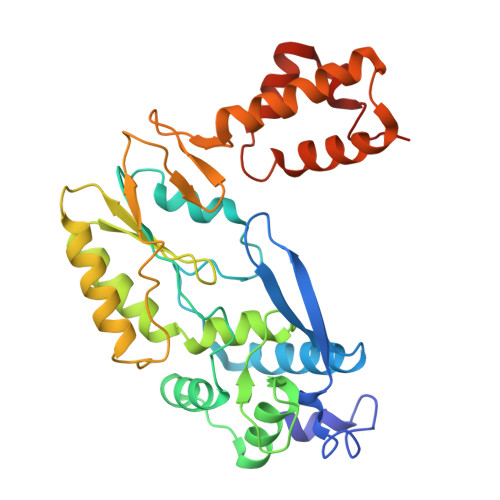
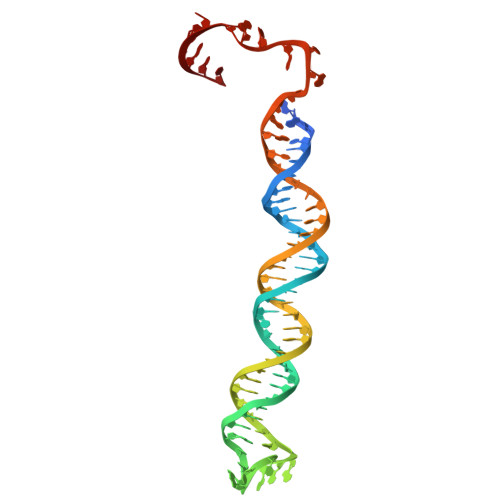
Disassembly activates Retron-Septu for antiphage defense.
Wang, C., Rish, A.D., Armbruster, E.G., Xie, J., Loveland, A.B., Shen, Z., Gu, B., Korostelev, A.A., Pogliano, J., Fu, T.M.(2025) Science 389: eadv3344-eadv3344
- PubMed: 40504952
- DOI: https://doi.org/10.1126/science.adv3344
- Primary Citation of Related Structures:
9E8Z, 9E90, 9E91, 9O4A - PubMed Abstract:
Retrons are antiphage defense systems that produce multicopy single-stranded DNA (msDNA) and hold promises for genome engineering. However, the mechanisms of defense remain unclear. The Retron-Septu system uniquely integrates retron and Septu antiphage defenses. Cryo-electron microscopy structures reveal asymmetric nucleoprotein complexes comprising a reverse transcriptase (RT), msDNA (a hybrid of msdDNA and msrRNA), and two PtuAB copies. msdDNA and msrRNA are essential for assembling this complex, with msrRNA adopting a conserved lariat-like structure that regulates reverse transcription. Notably, the assembled Retron-Septu complex is inactive, with msdDNA occupying the PtuA DNA-binding site. Activation occurs upon disassembly, releasing PtuAB, which degrades single-stranded DNA to restrict phage replication. This "arrest-and-release" mechanism underscores the dynamic regulatory roles of msDNA, advancing our understanding of antiphage defense strategies.
- Department of Pathology, UMass Chan Medical School, Worcester, MA, USA.
Organizational Affiliation: