Complex of Human MIRO1 and TRAK1 Binding Site-2 (L570-R613)
Ravitch, E.E., Baltrusaitis, E.E., Perez, T.P., Barrie, K.R., Fenton, A.R., Holbaur, E.L.F., Dominguez, R.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Mitochondrial Rho GTPase 1 | 618 | Homo sapiens | Mutation(s): 0 Gene Names: RHOT1, ARHT1 EC: 3.6.5 | 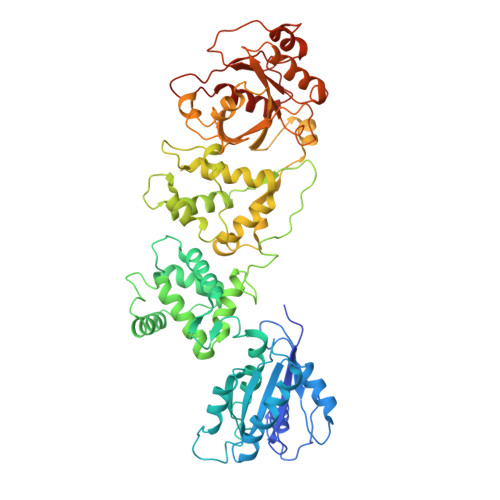 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q8IXI2 (Homo sapiens) Explore Q8IXI2 Go to UniProtKB: Q8IXI2 | |||||
PHAROS: Q8IXI2 GTEx: ENSG00000126858 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8IXI2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Trafficking kinesin protein 1 | 59 | Homo sapiens | Mutation(s): 0 Gene Names: TRAK1 | 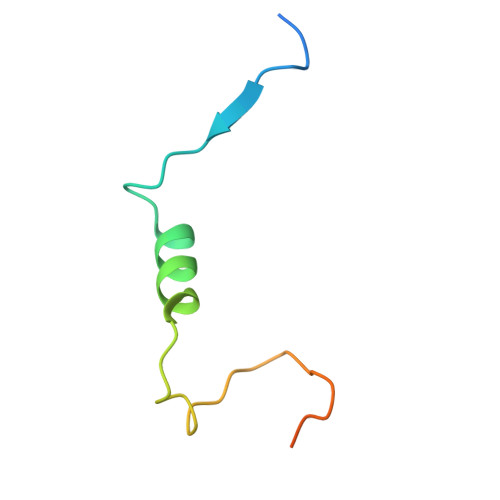 | |
UniProt | |||||
Find proteins for A0A8C9AJS8 (Prolemur simus) Explore A0A8C9AJS8 Go to UniProtKB: A0A8C9AJS8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8C9AJS8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GTP Query on GTP | E [auth A], G [auth A], J [auth C], L [auth C] | GUANOSINE-5'-TRIPHOSPHATE C10 H16 N5 O14 P3 XKMLYUALXHKNFT-UUOKFMHZSA-N |  | ||
| CA Query on CA | H [auth A], I [auth A], M [auth C], N [auth C] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| MG Query on MG | F [auth A], K [auth C] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | RM1 GM136511 |