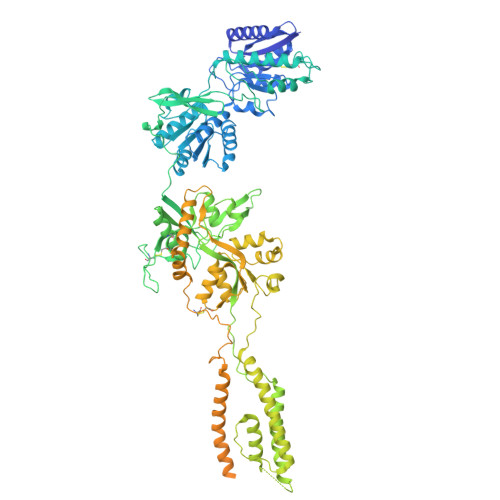
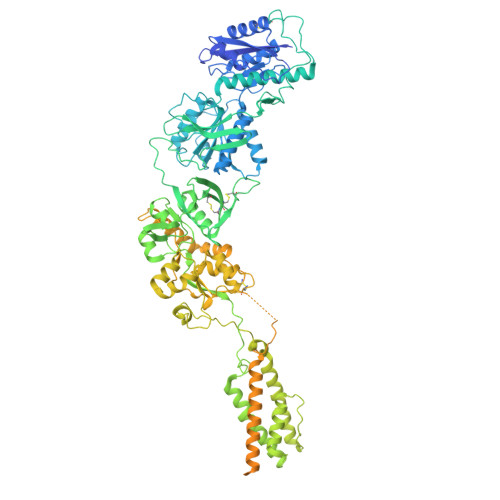
Cryo-EM snapshots of NMDA receptor activation illuminate sequential rearrangements.
Abbott, J.A., Kim, J., Liu, B., Popescu, G.K., Gouaux, E., Jalali-Yazdi, F.(2025) Sci Adv 11: eadx4647-eadx4647
- PubMed: 40991709
- DOI: https://doi.org/10.1126/sciadv.adx4647
- Primary Citation of Related Structures:
9C7C, 9C7E, 9C7P, 9C7Q, 9C7R - PubMed Abstract:
Canonical N -methyl-d-aspartate receptors (NMDARs) are glutamate-gated ion channels with critical roles in the development and function of the nervous system. The excitatory currents they produce reflect stochastic transitions between multiple agonist-bound closed- and open-pore states. We leveraged the intrinsically high open probability ( P o ) of NMDARs composed of GluN1 and GluN2A subunits, together with judiciously chosen mutants and ligands, to achieve conditions in which receptors had a P o near unity. Using single-particle cryo-electron microscopy (cryo-EM), we captured three activated receptor states, each with distinct conformations of the gate-forming M3 helices. Separately, we carried out single-channel electrophysiology, together with statistical modeling, to relate the cryo-EM structures to the gating reaction. NMDAR channel opening involves bending of the pore-forming M3 helices to produce a transient open-channel conformation, subsequently stabilized by new interactions between the D2-M3 linkers with the pre-M1 helices and the pre-M4 loops, to yield the stable open channel.
- Department of Biochemistry, Jacobs School of Medicine and Biomedical Sciences, University at Buffalo, SUNY, Buffalo, NY 14203, USA.
Organizational Affiliation: